Abstract
Evolutionary survival of a species is largely a function of its reproductive fitness. In mammals, a sparsely populated and widely dispersed network of hypothalamic neurons, the gonadotropin-releasing hormone (GnRH) neurons, serve as the pilot light of reproduction via coordinated secretion of GnRH. Since it first description, human GnRH deficiency has been recognized both clinically and genetically as a heterogeneous disease. A spectrum of different reproductive phenotypes comprised of congenital GnRH deficiency with anosmia (Kallmann syndrome), congenital GnRH deficiency with normal olfaction (normosmic idiopathic hypogonadotropic hypogonadism), and adult-onset hypogonadotropic hypogonadism has been described. In the last two decades, several genes and pathways which govern GnRH ontogeny have been discovered by studying humans with GnRH deficiency. More importantly, detailed study of these patients has highlighted the emerging theme of oligogenicity and genotypic synergism, and also expanded the phenotypic diversity with the documentation of reversal of GnRH deficiency later in adulthood in some patients. The underlying genetic defect has also helped understand the associated nonreproductive phenotypes seen in some of these patients. These insights now provide practicing clinicians with targeted genetic diagnostic strategies and also impact on clinical management.
Introduction
A unique neural network within the hypothalamus consisting of approximately 1,500 neurons termed the parvocellular system initiates and maintains reproductive function in mammals [1,2]. It accomplishes this task by coordinating the synthesis and pulsatile secretion of a single neuroendocrine decapeptide, gonadotropin-releasing hormone (GnRH), from this neural network. In humans, this GnRH neuronal network is fully active resulting in adult levels of activity of the hypothalamic-pituitary-gonadal axis during the late stages of gestation and the early neonatal period. It then becomes quiescent during childhood until its full reactivation at puberty [3,4]. These GnRH neurons reside predominantly in the medial preoptic area, project their axonal processes into the median eminence of the hypothalamus, and secrete GnRH into the hypophyseal portal circulation in a coordinated and pulsatile manner [5,6,7]. Once at its cognate pituitary receptor on the gonadotropes, GnRH stimulates both the synthesis and secretion of the two pituitary gonadotropic hormones: luteinizing hormone (LH) and follicle-stimulating hormone that, in turn, stimulate steroidogenesis and govern the maturation and function of the germ cells in the gonads in both sexes.
Remarkably, despite the critical evolutionary role of the reproductive system in survival of the species, human reproductive activity is invested in a single gene encoding GnRH and this unique network of hypothalamic neurons. Considering the ever-changing environmental demands that have threatened survival of mammals over the millennia, it is likely that a multi-tiered, overlapping network of genetic and cellular pathways has evolved to regulate the secretion of GnRH. The existence of such a complex yet fascinating biological system ‘upstream’ of GnRH secretion has been hinted at by the study of humans presenting with GnRH deficiency. These insights have been enabled by combining careful clinical investigations in these patients with the new powerful tools of developmental and molecular biology, genetics, animal models, and structural biology. Integration of these various tools and investigative approaches has allowed several groups around the world to begin to dissect genetic components of this unique neuroendocrine network and begin to define a functional map of the ontogeny of the GnRH neurons. This review attempts to put into perspective the evolving genetic architecture and molecular pathways governing the ontogeny of GnRH neurons as elucidated by the study of humans with GnRH deficiency, a truly prismatic disorder.
GnRH Neuronal Development
The first major advance that provided insight into the complexities of the developmental biology of the GnRH neurons came from investigations in rodents where GnRH neurons were demonstrated to originate outside the central nervous system (CNS) in the medial olfactory placode during embryological development [8,9,10]. From such humble origins where these neurons first specify their biological fate, committed GnRH neurons migrate from the olfactory placode along the olfactory epithelium to the olfactory bulb and then, via a structural scaffolding created by the olfactory bulb and tract, ultimately migrate to take up their final residence over a widely dispersed network within the medial preoptic area of the hypothalamus [11,12] (fig. 1a, b). These precise anatomic locations presumably reflect their need to determine and hence monitor a wide variety of sensory systems and other neuronal systems that provide data to the reproductive system required for survival (fig. 1b). These include body weight and nutritional status; circadian and light/dark cycles; stress and adrenal function; olfaction which helps locate predators as well as potential mating options; and sight and hearing of other environmental variables that might threaten the species. The GnRH neurons integrate these complex signals and determine whether the timing and environment are optimal to support full capability of the reproductive system by both sex steroid secretion and fertility or whether the optimal reproductive status is to silence and suppress both. The fine balance of timing of these two different settings on the GnRH scale then determines the ability of the individual and species to adapt and survive various environmental threats.
GnRH migration. a Migratory route of GnRH neurons in the embryonic mouse brain. GnRH neurons (black dots) originate in the medial wall of the olfactory placode, migrate along the olfactory axons and enter the brain perforating the cribriform plate. They then migrate to reach their final destination in the preoptic area (poa) of the hypothalamus. GnRH neurons on embryonic day 11 are seen in the vomeronasal organ (vno) and medial wall of the olfactory placode. In the 16-day-old fetal mouse brain, most of the GnRH neurons have reached the hypothalamus. gt = Ganglion terminale; ob = olfactory bulb. Reproduced with permission from Schwanzel-Fukuda and Pfaff [8]. b Cartoon of GnRH migration in humans. GnRH neurons originate in the embryonic olfactory placode and migrate along olfactory axons, penetrate the cribriform plate and migrate across the olfactory bulb, aided by various chemoattractive and repulsion factors and eventually reach the preoptic area of the hypothalamus, where they synchronize pulsatile GnRH secretion. Kisspeptin (KISS1) and the tachykinin (TAC) neurons serve as upstream modulators of GnRH secretion. Upon GnRH stimulation, the pituitary (PIT) secretes LH and follicle-stimulating hormone (FSH), which in turn regulate gonadal steroidogenesis and gametogenesis.
GnRH migration. a Migratory route of GnRH neurons in the embryonic mouse brain. GnRH neurons (black dots) originate in the medial wall of the olfactory placode, migrate along the olfactory axons and enter the brain perforating the cribriform plate. They then migrate to reach their final destination in the preoptic area (poa) of the hypothalamus. GnRH neurons on embryonic day 11 are seen in the vomeronasal organ (vno) and medial wall of the olfactory placode. In the 16-day-old fetal mouse brain, most of the GnRH neurons have reached the hypothalamus. gt = Ganglion terminale; ob = olfactory bulb. Reproduced with permission from Schwanzel-Fukuda and Pfaff [8]. b Cartoon of GnRH migration in humans. GnRH neurons originate in the embryonic olfactory placode and migrate along olfactory axons, penetrate the cribriform plate and migrate across the olfactory bulb, aided by various chemoattractive and repulsion factors and eventually reach the preoptic area of the hypothalamus, where they synchronize pulsatile GnRH secretion. Kisspeptin (KISS1) and the tachykinin (TAC) neurons serve as upstream modulators of GnRH secretion. Upon GnRH stimulation, the pituitary (PIT) secretes LH and follicle-stimulating hormone (FSH), which in turn regulate gonadal steroidogenesis and gametogenesis.
Once in place, these GnRH neurons extend axonal projections to the median eminence where they coordinate their secretion of GnRH in a pulsatile pattern of secretion that is absolutely required by the gonadotrope to evoke physiologic gonadotropin secretion [13] and gonadal function (fig. 1b). These GnRH migration studies are in line with the previously documented observation that when normal fetal GnRH neurons were transplanted into the third ventricles of GnRH-deficient adult female hypogonadal mice (hpg mice), transplanted GnRH neurons proved capable of projecting GnRH-containing processes to the median eminence in the host [14], initiating pulsatile LH secretion [15], and restoring limited reproductive competence [14] to those GnRH-deficient animals. The precise factors guiding this peripatetic journey of the GnRH neurons are unknown but experimental evidence supports involvement of a wide variety of genetic and biochemical cues including axonal guidance molecules, cell adhesion molecules, and various transcription and growth factors [16].
Human GnRH Deficiency: A Clinically and Genetically Heterogeneous Disease Model
Reactivation of the GnRH pulse generator during adolescence heralds the onset of puberty (fig. 2a). In boys, pubertal onset is marked by testicular enlargement, which is followed by penile and pubic hair growth. In girls, the first sign of puberty is the onset of breast budding (thelarche), followed by pubic hair growth and menarche. Normal ages for puberty are 8–13 years in girls and 9–14 years in boys. Puberty is ‘delayed’ when the secondary sexual characteristics fail to develop beyond 2 standard deviations from the mean. This delay in the activation of the GnRH pulse generator resulting in delayed puberty is the mildest form of human GnRH deficiency (fig. 2b). On the contrary, premature initiation of the GnRH pulse generator results in central precocious puberty (fig. 2c).
GnRH pulsatile secretion in normal puberty and disorders of pubertal development. a Pulsatile secretion of GnRH is fully active during the early neonatal period (‘mini-puberty of infancy’), followed by quiescence during childhood, and reactivates in adolescence signalling normal puberty. b Delayed activation of the GnRH pulse generator results in delayed puberty. c Premature activation of the GnRH pulse generator in early childhood results in precocious puberty.
GnRH pulsatile secretion in normal puberty and disorders of pubertal development. a Pulsatile secretion of GnRH is fully active during the early neonatal period (‘mini-puberty of infancy’), followed by quiescence during childhood, and reactivates in adolescence signalling normal puberty. b Delayed activation of the GnRH pulse generator results in delayed puberty. c Premature activation of the GnRH pulse generator in early childhood results in precocious puberty.
In 1856, Maestre de San Juan [17] first documented the pathological association of hypogonadism and absence of the olfactory system. A more detailed description of the syndrome was reported in 1944 by Kallmann [18] using patients in three affected pedigrees with hypogonadism and anosmia. His observations were the first to hint at a broader spectrum of clinical defects and identify the familial nature in the clinical syndrome that was seen in both sexes and accompanied by multiple congenital anomalies. In 1954, de Morsier [19 ]first noted the link between hypogonadism and neuroanatomical defects including agenesis of the olfactory bulb and tract and other midline neuroanatomical defects and coined the term ‘olfacto-genital dysplasia’. Since then, hypogonadotropic hypogonadism accompanied by anosmia has been classically described as Kallmann syndrome (KS) (fig. 3a). Eventually, the discovery of GnRH in 1971 aided the linking of the etiology of KS to hypogonadotropic hypogonadism [20,21]. Although the variability of the pituitary gonadotropin response to a single bolus of GnRH initially failed to definitively confirm hypothalamic GnRH deficiency [22], subsequent normalization of the pituitary-gonadal axis in response to a physiologically defined regimen of exogenous GnRH administration in most of these patients confirmed the hypothalamic defect in these patients [23]. However, a pituitary defect in GnRH action was subsequently demonstrated by the discovery of mutations in GnRH receptor (GNRHR) in some patients with hypogonadotropic hypogonadism [24]. Hypogonadotropic hypogonadism is also known to occur in the presence of normal olfaction and is classified as normosmic idiopathic hypogonadotropic hypogonadism (nIHH) (fig. 3b). In this regard, it is noteworthy that even in Kallmann’s [18] initial report, 3 of the original 12 hypogonadal individuals had no obvious defect in olfaction.
KS and nIHH. a KS results from a developmental defect in olfactory bulb (OB) morphogenesis and GnRH neuronal migration resulting in failure of GnRH neurons to arrive at the preoptic area (POA) in the hypothalamus and failure of GnRH secretion. The GnRH neurons are arrested in their extracerebral route. Mutations in KAL1, PROK2, PROKR2, FGFR1, FGF8, NELF and CHD7 have been associated with KS. b nIHH results from either defective pulsatile secretion of GnRH or defective GnRH action at the pituitary. Mutations in GNRH1, KISS1R, GNRHR, PROK2, PROKR2, FGFR1, TAC3, TACR3, CHD7 and NROB1 genes have been associated with nIHH.
KS and nIHH. a KS results from a developmental defect in olfactory bulb (OB) morphogenesis and GnRH neuronal migration resulting in failure of GnRH neurons to arrive at the preoptic area (POA) in the hypothalamus and failure of GnRH secretion. The GnRH neurons are arrested in their extracerebral route. Mutations in KAL1, PROK2, PROKR2, FGFR1, FGF8, NELF and CHD7 have been associated with KS. b nIHH results from either defective pulsatile secretion of GnRH or defective GnRH action at the pituitary. Mutations in GNRH1, KISS1R, GNRHR, PROK2, PROKR2, FGFR1, TAC3, TACR3, CHD7 and NROB1 genes have been associated with nIHH.
Mutations in a number of different genes have now been linked with human GnRH deficiency, but only approximately 40% of the genetic heritability has been uncovered, with a number of genes yet to be discovered (table 1). However, the growing appreciation that both KS and nIHH can occur within a given family (i.e. with the same genetic milieu) demonstrated that both of these variants of GnRH deficiency are part of a continuous spectrum of clinical and genetic defects rather than single unique phenotypes. Those with anosmia presumably signal a joint involvement of the olfactory system and GnRH neurons in the pathophysiology of this disorder, whereas those with a normal sense of smell indicate processes occurring either before or after migration of GnRH neurons. In addition, several patients with KS and nIHH present with varying degrees of pubertal delay, with some presenting with complete absence of puberty suggesting complete GnRH deficiency (fig. 4a) and others with partial pubertal development indicating partial GnRH deficiency (fig. 4b). Several other additional phenotypes including isolated anosmia and delayed puberty also form part of this clinical spectrum [25,26,27,28]. Moreover, the presence of phenotypically normal individuals who carry a similar genetic burden as other affected family members has uncovered the critical role played by oligogenicity and genotypic synergism in the observed clinical phenotype [29,30].
Phenotypic heterogeneity in human GnRH deficiency. GnRH deficiency can either be complete (a) or partial (b). c Some patients develop GnRH deficiency in adulthood following normal GnRH activation in the neonatal period (AHH). d Occasionally, following complete or partial GnRH deficiency, GnRH pulse generator activates in adulthood (reversal of GnRH deficiency).
Phenotypic heterogeneity in human GnRH deficiency. GnRH deficiency can either be complete (a) or partial (b). c Some patients develop GnRH deficiency in adulthood following normal GnRH activation in the neonatal period (AHH). d Occasionally, following complete or partial GnRH deficiency, GnRH pulse generator activates in adulthood (reversal of GnRH deficiency).
Human GnRH deficiency had traditionally been thought to be a monogenic disorder with complete and permanent failure of sexual maturation due to silencing of the GnRH pulse generator. However, it has become increasingly clear that human GnRH deficiency is not merely a congenital arrest of GnRH neuronal development as was implied by the initial studies of KAL1- deleted patients (see below). The identification of adult-onset idiopathic hypogonadotropic hypogonadism (AHH) in men confirmed normal sexual development until the cessation of GnRH pulse generator function that occurs well into adulthood [28] (fig. 4c). Similarly, spontaneous reversals of well-established GnRH deficiency following long-term therapy with sex steroids has added yet another clinical variant suggesting a more complex pathophysiology than was initially thought [31] (fig. 4d).
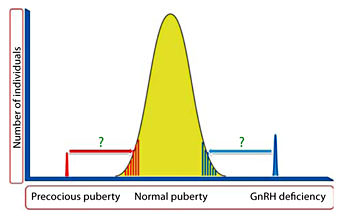
Thus, it is becoming increasingly clear that GnRH deficiency in humans represents a unique family of oligogenic disorders that present with a continuum of reproductive and non-reproductive phenotypes. These phenotypes belie both developmental and functional pathophysiological defects and in some cases can reverse completely in adulthood. Detailed clinical investigations of human GnRH deficiency aided by diverse basic research tools have proven to provide a prism into the fascinating complexities of GnRH deficiency and the ontogeny of this neuronal network. It is possible that the rare genetic variants that underlie human GnRH deficiency and possibly central precocious puberty may eventually explain the heritability of delayed puberty and early puberty seen within the normal population (fig. 5).
Genetics of puberty.The precise genetic basis of the extreme tails of normal puberty is unclear. The genetic variants causing GnRH deficiency and precocious puberty may possibly explain the heritability of early and delayed puberty in the normal population.
Genetics of puberty.The precise genetic basis of the extreme tails of normal puberty is unclear. The genetic variants causing GnRH deficiency and precocious puberty may possibly explain the heritability of early and delayed puberty in the normal population.
Discovery of KAL1
The first human gene responsible for KS was localized to the distal portion of the X chromosome by investigators studying a pedigree with 5 male patients with X-linked ichthyosis, hypogonadotropic hypogonadism and anosmia [32]. This finding was extended in a subsequent study of a male infant born with KS associated with ichthyosis, chrondroplasia punctata and choanal atresia and chromosomal analysis showed deletion of the Xp22.3 region of the X chromosome inherited from his unaffected mother [33]. The child subsequently died and an autopsy showed complete absence of olfactory bulbs and tracts and absence of cribriform plate perforations [33]. A second male child was then conceived by the same mother and amniocentesis revealed an identical defective X chromosome. Neuroanatomical studies of the aborted fetus at 19 weeks of gestation revealed that the GnRH neurons had developed, migrated from the olfactory placode towards the CNS, yet were arrested in their migration at the cribriform plate of the ethmoid bone, presumably since there were no olfactory axons to guide their transport further [34].
Eventually, the first KS gene, KAL1, was identified by positional cloning of the distal portion of the X chromosome (Xp22.3) simultaneously by two independent groups [35,36]. The KAL1 gene (fig. 6b) is comprised of 14 exons, is yet to be discovered in the mouse, and encodes a 680-amino acid secreted extracellular-matrix glycoprotein, anosmin-1 (fig. 6c). Anosmin-1 is a secreted multi-domain protein consisting of an N-terminal signal peptide followed by a cysteine-rich region (cys-box), a whey acidic protein (WAP)-like four disulfide core motif, four tandem fibronectin type III (FnIII) repeats and a C-terminal histidine-rich region [35,36].
KAL1 mutations in human GnRH deficiency. a Gene deletions (black lines) in KAL1 gene causing KS. b Schematic of exons of KAL1 gene. c Schematic of the anosmin-1 protein displaying known point mutations (in hexagons) causing KS. Anosmin-1, a multidomain protein, consists of a N-terminal signal peptide followed by a cysteine-rich region (CYS), a WAP-like four-disulfide core motif, four tandem FnIII repeats and a C-terminal histidine-rich region (H) (insertions, deletions, duplications and intronic changes not shown).
KAL1 mutations in human GnRH deficiency. a Gene deletions (black lines) in KAL1 gene causing KS. b Schematic of exons of KAL1 gene. c Schematic of the anosmin-1 protein displaying known point mutations (in hexagons) causing KS. Anosmin-1, a multidomain protein, consists of a N-terminal signal peptide followed by a cysteine-rich region (CYS), a WAP-like four-disulfide core motif, four tandem FnIII repeats and a C-terminal histidine-rich region (H) (insertions, deletions, duplications and intronic changes not shown).
Anosmin-1 is required for the formation of the olfactory guidance platform for GnRH neuronal migration, but its precise biological activity in achieving this function is unclear. Similarly, a wave of its expression in the kidney precedes the appearance of glomeruli during renal development. This may well explain why some KS patients with mutations in KAL1 have congenital defects of the kidney including complete renal agenesis. Human anosmin-1 shares significant homology with neural cell adhesion molecules that have fibronectin repeats [35,36]. Although the search for the rodent homologue of anosmin-1 has been elusive, known KAL1 orthologs in other species are highly conserved, especially at the WAP and FnIII domains [37]. The eight cysteine residues in the WAP domain of anosmin-1 which form four disulfide bonds are also highly conserved amongst other serine protease inhibitors that are members of this WAP protein family [38]. Since anosmin-1 is a secreted protein, it is thought to act locally at the cell surface via its interaction with the cell membrane that depends largely on the binding of one of the 4 FnIII domains to heparan sulfate proteoglycans [38,39]. Expression of anosmin-1 is regionally and temporally restricted during human organogenesis [40]. Migration of immortalized rodent GnRH neurons is stimulated by human anosmin-1 [16]. Anosmin-1 immunoreactivity in human embryos has been shown to be present in the nasal placode as early as embryonic week 4.5 and its expression is seen in the presumptive olfactory bulb by the 5th embryonic week even prior to the olfactory nerve synaptogenesis with the olfactory bulb [40]. However, the absence of anosmin-1 in the olfactory epithelium or the extracerebral route of the olfactory axonal elongation suggests that the role of anosmin-1 may primarily rest in the intracerebral olfactory axonal elongation and subsequent GnRH neuronal migration along this course. This suggestion is further supported by the arrest of GnRH neuronal migration at the cribriform plate in the KS fetus lacking KAL1 [34]. Taken together, these findings suggest that the biological role of anosmin-1 in the presumptive olfactory bulb may be critical to attracting olfactory axons towards the forebrain. In the absence of anosmin-1, this trajectory is lost and GnRH neurons thus fail to reach the olfactory bulb initially and subsequently the hypothalamus [41]. Apart from its role in olfactory axonal attraction, it is likely anosmin-1 may independently play a role in olfactory bulb morphogenesis per se and/or neurite outgrowth beyond the olfactory bulb through the lateral olfactory tract [42].
Despite the critical developmental role played by anosmin-1, deletions in KAL1 (fig. 6a) and mutations in KAL1 (fig. 6c) only account for up to 10–14% of familial KS [43] and 8–11% of sporadic KS [44] patients. A wide spectrum of mutations spanning the entire coding sequence of the KAL1 gene have been reported in X-linked KS subjects [45]. Many of the point mutations cluster around the FnIII domain [46], confirming the key molecular role played by this domain in protein activity [39,47]. In human subjects with KAL1 mutations, several nonreproductive features, including unilateral renal agenesis, synkinesia or mirror movements, hearing loss, midline facial defects, and bony abnormalities are variably seen [44,48]. In keeping with the clinical signatures, the wider developmental roles of anosmin-1 are strongly supported by the detection of the protein in the extracellular matrices of various nonreproductive tissues such as bronchial tubes, mesonephric tubules, ureteric bud, digestive tract, large blood vessels, and inner ear [40].
KISS1R (GPR54)
The relative rarity of human GnRH deficiency, its multiple inheritance patterns, its associated reduced fertility, and consequent small pedigree sizes have rendered traditional linkage analysis less effective in aiding novel gene discovery in this phenotype. However, in 2003, using linkage analysis in inbred families, two groups independently identified GPR54, a G protein-coupled receptor, and its cognate ligand, kisspeptin, to be upstream gatekeepers of GnRH neurons [49,50]. Coupled with complementary mouse genetics and in vitro confirmation of loss of biological activity of the mutant receptor protein, GPR54 was implicated as a key regulator of puberty [50] and a number of mutations in this receptor have now been described in nIHH patients (fig. 7a, b). GPR54, which has recently been redesignated as KISS1R, is a member of the rhodopsin family of G protein-coupled receptors with sequence homologies to the members of the galanin receptor family [51]. Kisspeptins, an overlapping family of RF-amide peptides, serve as endogenous ligands for KISS1R and are potent stimulators of gonadotropin release in all mammalian species studied to date [51,52]. Kisspeptin is coded by the KISS1 gene and its proteolytic processing results in different-length kisspeptins, all of those which retain the amidated carboxy-terminal decapeptide are potent KISS1R agonists [51,53]. The longest kisspeptin, kisspeptin-1, is a 54-amino acid peptide and was initially termed metastin due to its ability to inhibit tumor metastasis [53]. So far, KISS1 mutations causing human GnRH deficiency have not been reported.
KISS1R and human GnRH deficiency. a Schematic of the predicted KISS1R protein displaying known mutations causing nIHH in humans. b Dose-response curves for the ligand-stimulated production of inositol phosphate in KISS1R mutant constructs, corrected for protein content. i Curve for the L148S mutation (three independent experiments, each performed in triplicate). ii Curve for the R331X mutation (two independent experiments, each performed in triplicate). iii Curve for the X399R polyA stop mutation (two independent experiments, each performed in triplicate). The percentages on the y-axis represent the percentages of the maximal stimulation for each GPR54 construct. iv The relative quantification of the wild-type and mutant GPR54 allele expression in lymphoblastoid cell lines as measured by quantitative RT-PCR. Reproduced with permission from Seminara et al. [50].
KISS1R and human GnRH deficiency. a Schematic of the predicted KISS1R protein displaying known mutations causing nIHH in humans. b Dose-response curves for the ligand-stimulated production of inositol phosphate in KISS1R mutant constructs, corrected for protein content. i Curve for the L148S mutation (three independent experiments, each performed in triplicate). ii Curve for the R331X mutation (two independent experiments, each performed in triplicate). iii Curve for the X399R polyA stop mutation (two independent experiments, each performed in triplicate). The percentages on the y-axis represent the percentages of the maximal stimulation for each GPR54 construct. iv The relative quantification of the wild-type and mutant GPR54 allele expression in lymphoblastoid cell lines as measured by quantitative RT-PCR. Reproduced with permission from Seminara et al. [50].
Review of human and mouse KISS1R investigations have provided fascinating insights into the role of KISS1/KISS1R biology in human reproduction. Both KISS1–/– and KISS1R–/– mice are phenocopies of nIHH and interestingly show normal GnRH content in the hypothalamus, providing the first indication that mutations in KISS1R do not affect GnRH neuronal migration or GnRH synthesis but rather GnRH release [50,54]. Several key observations have emerged from the study of human GnRH-deficient subjects with KISS1R mutations. Neuroendocrine profiling of probands with KISS1R mutations has generally shown dampened but present low-amplitude LH pulses [50,55] suggesting some degree of endogenous GnRH secretion that is synchronized but reduced in pulse amplitude. An African-American proband with a compound heterozygous mutation in KISS1R also showed a striking leftward shift in his LH dose-response relationship to exogenous GnRH. This observation suggests some degree of endogenous pituitary priming by intact but dampened GnRH pulsatility [50]. In one published report of a male with KISS1R mutation who presented with cryptorchidism and micropenis in infancy, neuroendocrine evaluation at 2 months of age revealed undetectable gonadotropins also suggesting a role for the KISS1/KISS1R system in the ‘mini-puberty’ of infancy [56]. Some patients with KISS1R mutations are able to undergo folliculogenesis, spermatogenesis and successful pregnancies following therapy with exogenous GnRH, suggesting some degree of intact pituitary gonoadotropin function with no significant primary gonadal defects or defects in placentation in these individuals [57].
Although a variety of diverse phenotypes is often seen in human GnRH-deficient subjects, nonreproductive phentoypes have yet to be described in subjects with KISS1R mutations. However, both KISS1 and KISS1R expression are not restricted to the hypothalamic-pituitary-gonadal axis and both the ligand and receptor expression are reported elsewhere in the body such as the adrenal, placenta, and pancreatic islet beta cells [53]. Moreover, both KISS1 and KISS1R were implicated as metastasis suppressors in cells from breast, melanoma, and pancreatic cancers [53]. The KISS1/KISS1R pathway has also been implicated in hypothalamic control of energy balance [58], glucose-induced insulin secretion in the pancreas [59] and has been postulated to be a potential integrator of the gonadal hormonal and circadian signals and relay to the GnRH neurons [60].
Although mutations in the KISS1/KISS1R pathway do not seem to be a significant contributor to human GnRH deficiency (<5%), their relative rarity probably suggests an evolutionarily critical role in reproduction and species propagation that might well have undergone negative selection. The recently reported documentation of a ligand-independent gain-of-function mutation in GPR54 in a girl with idiopathic central precocious puberty demonstrates the crucial role of this system in the maturation of the reproductive axis [61]. Furthermore, constant infusion of the terminal decapeptide of kisspeptin to rhesus monkeys is capable of inducing a homologous desensitization of the KISS1R receptor despite retained GnRH responsiveness at the level of the gonadotrope [62]. Thus, the discovery of this previously unsuspected pathway in human reproduction that has now been discovered to represent a key control point of GnRH at sexual maturation in all mammals studied to date has ignited a burst of basic and clinical research into the KISS1/KISS1R axis in mammalian physiology. This system offers a further approach to therapeutically modulate human GnRH function and potentially translate the biology of this pathway into wider clinical applications across diverse reproductive phenotypes.
Fibroblast Growth Factor Signalling and GnRH Ontogeny
The family of 23 fibroblast growth factors (FGFs) and their 4 cognate receptors (FGFRs) have multiple roles in central nervous and skeletal system development and include brain patterning, branching morphogenesis and limb development [63]. In 2 individuals with a contiguous gene syndrome with a dominant form of KS, Dodé et al. [64] first identified an overlapping interstitial deletion at chromosome 8p11–p12 using fluorescent in situ hybridization and reported loss-of-function mutations in FGFR1 as the underlying cause of KS. This finding not only convincingly demonstrated FGFR1 to be KAL2 but also explained the long-standing paradox of patients with both GnRH deficiency and hereditary spherocytosis due to ankyrin deficiency. Both these genes are located in a region quite close to the GnRH gene itself which had not been identified as a cause of GnRH deficiency until recently. Again, it was GnRH-deficient patients with mutations in this gene that heralded the recognition of the relatively crucial role of the FGFs in mammalian reproduction.
There are 23 mammalian FGFs that act by binding and activating one of the 4 FGFR family of tyrosine kinase receptors in a heparan sulfate proteoglycan-dependent manner [63]. Four FGFR genes (FGFR1–FGFR4) encode their respective receptors. Each of these 4 FGFRs consists of three extracellular immunoglobulin domains (D1–D3), a single-pass transmembrane domain and a cytoplasmic tyrosine kinase domain [65] (fig. 8a). An ‘acidic box’ consisting of a serine-rich sequence present in the D1–D2 linker and in conjunction with the D1 domain is thought to mediate autoinhibition [63]. Within the first half of D2 there is a site that binds the FGFR coreceptor, heparan sulfate proteoaminoglycan. The D2 and D3 domains are responsible for ligand binding and specificity. Alternative splicing of the FGFR genes produces 7 principal isoforms and each isoform exhibits differential ligand binding specificities [66]. After binding of their cognate ligand(s) and heparan sulfate proteoglycan(s), the FGFRs dimerize, resulting in autophosphorylation and stimulation of the protein tyrosine kinase activity. This activation step is followed by the stimulation of an extensive intracellular signalling cascade involving various signal proteins, coreceptor proteins and various second messengers that are described in detail elsewhere [63]. This FGFR1 signalling activity is tightly regulated and in quiescent cells, the acidic box domain of these receptors is thought to bind to the positively charged heparan sulfate binding site and result in a ‘closed’ inactive state that is in equilibrium with the ‘open’ active one. Upon ligand availability, the equilibrium shifts towards the ‘open’ state.
FGF pathway in human GnRH deficiency. a Schematic of FGFR1 protein displaying mutations causing KS (blue) and nIHH (black). The extracellular domain of FGFR1 contains a signal peptide (SP), three extracellular immunoglobulin-like (D1, D2, and D3) domains, followed by the TM, and an intracellular domain comprising two tyrosine kinase subdomains (PTK). The acidic box (AB) and the docking protein FRS2 domain in the extracellular domain and intracellular domain, respectively, are specific features of FGF receptors. b Genomic structure and differential splicing of the human FGF8 gene. i Structure of the FGF8 gene. Boxes denote exons; lines denote introns. ii Schematic of the four FGF8 isoforms identified in humans with mutations identified to date are indicated by arrows and numbered according to the FGF8f and FGF8b protein isoforms (* homozygous mutations). Reproduced from Falardeau et al. [71].
FGF pathway in human GnRH deficiency. a Schematic of FGFR1 protein displaying mutations causing KS (blue) and nIHH (black). The extracellular domain of FGFR1 contains a signal peptide (SP), three extracellular immunoglobulin-like (D1, D2, and D3) domains, followed by the TM, and an intracellular domain comprising two tyrosine kinase subdomains (PTK). The acidic box (AB) and the docking protein FRS2 domain in the extracellular domain and intracellular domain, respectively, are specific features of FGF receptors. b Genomic structure and differential splicing of the human FGF8 gene. i Structure of the FGF8 gene. Boxes denote exons; lines denote introns. ii Schematic of the four FGF8 isoforms identified in humans with mutations identified to date are indicated by arrows and numbered according to the FGF8f and FGF8b protein isoforms (* homozygous mutations). Reproduced from Falardeau et al. [71].
A large number of individual point mutations and deletions of the FGFR1 gene have now been reported in subjects with both KS [26,42] and nIHH [67] (fig. 8a). These mutations span all the functional domains of the FGFR1 receptor and, in contrast to KAL1, account for up to 10% of each KS and nIHH patient series [67,68]. Structural and biochemical studies using recombinant FGFR1 proteins to study the structure-activity relationships of these mutations have been particularly informative. Mutations in the D2 and D3 loops of the receptor were shown to cause misfolding and impaired cell-surface expression of the receptor and/or ligand-receptor interaction and binding [27,67]. Mutations in the cytoplasmic tyrosine kinase domain appear to cause structural perturbations and reduce catalytic activity of this domain [67]. At least 11 different FGFs can activate FGFR1 [69]. However, the specific ligand implicated in GnRH neuron ontogeny was unknown until a proband with KS and cleft palate with a L342S mutation in the FGFR1c gene demonstrated that this mutation results in a selective and dramatic loss of binding affinity of the FGF8b isoform to the FGFR1 receptor. Integrating this observation with the described overlapping patterns of FGF8 and FGFR1 expression in the brain and defective olfactory bulb neurogenesis and loss of fate specification of GnRH neurons seen in FGF8 hypomorphic mice [70], a candidate gene approach revealed six FGF8 gene mutations in GnRH-deficient unrelated probands [71] (fig. 8b). Two of the probands also carried additional FGFR1 mutations confirming oligogenicity in the FGF8 signalling pathway in human GnRH deficiency. The structural and functional impact of these FGF8 mutations were confirmed by a crystal structure analysis of the FGF8b/FGFR2c/heparin model and reduced FGF8 function in vitro [71].
These findings of FGFR1 and FGF8 mutations in both KS and nIHH add a new dimension to the complexity of GnRH ontogeny. Impaired FGF signalling can clearly result in loss of fate specification of GnRH neurons and/or GnRH migratory defects with impaired olfactory epithelium/bulb neurogenesis resulting in the KS phenotype and through alternative mechanisms affect GnRH neuronal survival beyond olfactory neurogenesis resulting in a nIHH phenotype. The effect of FGF signalling on olfactory bulb neurogenesis may, in fact, relate to the association between anosmin and FGFR1 proteins. Both anosmin-1 and FGFR1 bind to heparan sulfate proteoglycans. Anosmin-1 has been shown in vitro to enhance the mitogenic effect of the FGFR1c isoform on lymphoid cells in the presence of FGF2 [72]. These above associations have led to the postulation that X-linked KAL1 mutations may be secondary to a FGF signalling defect on GnRH neuronal development [73]. FGF signalling has previously been implicated in GnRH neurite outgrowth and GnRH axonal guidance to the median eminence and this observation suggests a chemoattractant role for FGF8 [74]. The importance of the FGF signalling pathway in GnRH neuron ontogeny is further supported by the murine models with targeted ablation of FGFR1 in the telencephalon [75] and FGF8 hypomorphic mice [70], both of which lack olfactory bulbs. In addition, mice expressing a dominant-negative form of FGFR1 in their GnRH neurons and FGF8 hypomorphic mice exhibit reduced or absent GnRH neurons in the hypothalamus [76]. Since complete reversal of GnRH deficiency has been documented in subjects carrying mutations in FGF8 and FGFR1, there may well be a role for FGF signalling in GnRH neuronal survival and apoptosis pathways [26,31,71,77].
Prokineticin 2/Prokineticin Receptor 2 and GnRH Ontogeny
Targeted gene deletions in animals often unearth unsuspected phenotypes. The prokineticin system is a prime example of this phenomenon as it was initially studied because of its importance in gastrointestinal function. Prokineticins (PROK1 and 2) are the mammalian protein orthologs of proteins discovered in the black mamba snake’s venomous secretions [78]. PROKR1 and PROKR2 are their respective receptors. These two receptors differ little in their ligand preference since they are >85% homologous in their DNA sequence. In fact, they were originally thought to be two different isoforms of the same receptor (i.e. GPR73a and GPR73b) until it was appreciated that they were encoded by two different genes. However, unanticipated hypogonadotropism occurred in murine deletions of the PROK2 pathway. While PROK1–/–[79] and PROKR1–/–[80] animals lack any CNS phenotypes, PROK2–/– [79,81] and PROKR2–/–[80] mice show olfactory bulb anomalies (fig. 9b), decreased GnRH neuronal migration to the hypothalamus, and hypogonadotropic hypogonadism responsive to exogenous gonadotropins. Although the olfactory bulb morphogenesis is completely disrupted in PROKR2–/– mice, in 50% of PROK2–/– mice, some olfactory bulb morphogenesis is preserved, suggesting that although PROK2 plays an important role in neuronal migration, several other key molecules/gene products are likely to be involved in the orchestration of GnRH neuronal migration and olfactory bulb development. Likewise, the evolving role of oligogenicity in GnRH deficiency suggests that although individual gene products play a critical role in determining the reproductive phenotype, interaction and synergy between various genes make a cumulative contribution to the eventual clinical phenotype.
Prokineticin 2 pathway and human GnRH deficiency. a Schematic of the predicted PROKR2 protein displaying mutations causing KS (yellow boxes) and nIHH (white boxes). b Morphological examination of wild-type PROK2+/+ and PROK2–/– mice showing hypoplasia of olfactory bulbs (red circle). Reproduced with permission from Ng et al. [79]. c In vitro activity of a selection of PROKR2 mutants in an intracellular Ca2+ assay. Reproduced with permission from Cole et al. [90].
Prokineticin 2 pathway and human GnRH deficiency. a Schematic of the predicted PROKR2 protein displaying mutations causing KS (yellow boxes) and nIHH (white boxes). b Morphological examination of wild-type PROK2+/+ and PROK2–/– mice showing hypoplasia of olfactory bulbs (red circle). Reproduced with permission from Ng et al. [79]. c In vitro activity of a selection of PROKR2 mutants in an intracellular Ca2+ assay. Reproduced with permission from Cole et al. [90].
The ligands, PROK1 (85 amino acids) and PROK2 (81 amino acids), are multifunctional secreted proteins with only 44% shared homology [78]. However, both ligands share 10 highly conserved cysteines that presumably pair in 5 bridges and have an identical basic N-terminal amino acid sequence, AVITGA, that is essential for their biological activity [82]. PROKR1 and PROKR2 are G-protein-coupled receptors whose major biological differences lie more in their differential pattern of expression than in their ability to distinguish between ligands. Previously, the ‘prokineticin system’ drew attention because of its broadly diverse biologic functions including its ability to stimulate gastrointestinal motility [83], angiogenesis [84], hematopoiesis [85] and its effect on pain modulation [86]. In contrast to the more widely expressed PROKR1, the ligand PROK2, and its cognate receptor, PROKR2, have a unique expression profile within the CNS including olfactory system, arcuate nucleus, suprachiasmatic nuclei (SCN) and median eminence [79,80]. Yet another fascinating feature of PROK2 is its known function as a chemoattractant for neural progenitor cells that ultimately populate the olfactory bulb and assist in its dynamic function during life [79]. Within the CNS, therefore, the combined anatomic expression pattern of PROK2/PROKR2 matches with systems that coordinate smell, reproduction, temperature, and circadian rhythms – all critical factors for successful evolution of species. Sensing animals of the opposite sex ready for breeding, determining internal reproductive readiness, and facilitating sensing of light/dark changes are all functions essential for reproductive ‘fitness’.
Because of the strong expression of PROK2/PROKR2 in the SCN and of PROKR2 in the paraventricular nucleus, a key SCN output target for the endocrine system, this signalling system is a key candidate for a potential link between the reproductive and circadian systems. In addition, the induction of PROK2’s expression by the clock genes, BMAL1 and CLOCK [79], support the notion that this ligand/receptor pair are strong candidates to serve as a potential second messenger system from the SCN’s master clock to the more peripheral and diverse circadian endocrine rhythms such as the reproductive system. Furthermore, both PROK2–/– and PROKR2–/– animals exhibit disruptions of some of their circadian rhythms including abnormal thermogenesis, increased nocturnal physical activity, impaired circadian cortisol, and abnormal glucose regulation [87,88,89].
In keeping with these observations in PROK2–/– and PROKR2–/– mice, humans with loss-of-function mutations in both PROK2 and PROKR2 have been documented to have both KS and nIHH [81,90,91] (fig. 9a, c). Similar to the subjects with FGF8/FGFR1 mutations, considerable phenotypic variability is evident within family members and some asymptomatic individuals carry identical mutations to other affected individuals within sibships. This variable expressivity and incomplete penetrance that are now emerging in several genes causing GnRH deficiency strongly suggest the existence of a digenic/oligogenic phenomenon as previously documented [90,91]. Subjects with mutations in the PROK2/PROKR2 system also have been documented to undergo reversal of their hypogonadotropism following treatment with sex steroids. This observation suggests that some degree of GnRH neuronal plasticity in adulthood, possibly modulated by sex steroids, is responsible for their improvement [90]. Any connection between the integrity of the circadian system in the SCN and the GnRH system is yet to be determined in the humans. However, several nonreproductive phenotypes are seen with prokineticin mutations including sleep disorders and obesity [90,91]. However, detailed circadian assessment in patients with mutations in this system will be required to ascertain if prokineticin is a major output molecule linking the circadian clock and reproduction.
GNRH1 and GNRHR
The human GNRH1 gene is located at 8p21–8p11.2, consists of 4 exons and encodes the preprohormone that is ultimately processed to produce GnRH decapeptide [92]. Mutation in GNRH1 is an obvious candidate as an etiology for human GnRH deficiency and in keeping with this, in the hypogonadal (hpg) mouse model, a homozygous deletion of the GNRH1 gene resulted in hypogonadotropic hypogonadism [14]. Although human GNRH1 mutations have been elusive for many years, two independent groups have recently described homozygous frameshift GNRH1 mutations in patients with GnRH deficiency [93,94]. In both studies, consistent with the critical role played by GnRH, subjects with homozygous mutations in GNRH1 showed severe hypogonadism with affected males having microphallus. In addition to homozygous frameshift mutations, heterozygous rare sequence variants in GNRH1 have also been described in one of these studies [93]. As seen with other genes implicated in GnRH neuronal ontogeny, it is likely that oligogenicity and genotypic synergy with known/unknown genes may operate to produce the phenotype. No non-reproductive features were reported in these patients.
The amino acid sequence of GnRHR was first deduced for the mouse receptor cloned from the pituitary αT3 gonadotrope cell line [95]. The human GNRHR gene maps to chromosome 4q13.2–13.3 and is comprised of three exons that encode a 328-amino acid protein, with >85% homology within mammalian species, with near identity in the transmembrane domains. The GnRHR contains seven transmembrane (TM) domains, six of which alternate extra- and intracellular loops with an extracellular amino terminus. However, GnRHR is unique among the rhodopsin family of GPCRs in its lack of an intracellular carboxy terminus [96]. The extracellular domains and superficial regions of the TMs are involved in binding of GnRH, and the TMs are believed to be involved in receptor configuration and conformational change associated with signal propagation (receptor activation) [97]. These changes are thought to propagate into conformational changes in the intracellular domains involved in interacting with G proteins and other proteins for intracellular signal transduction [97]. GnRH binds to the GnRHR in a hairpin structure with the amino- and carboxy-terminal domains contributing mainly to receptor binding and activation [98].
Since the original reports of GNRHR mutations causing nIHH [99], a variety of inactivating mutations have been described. Most are missense mutations and a significant number of compound heterozygous changes are seen. The most common mutations occur in the first extracellular and third intracellular loops, although they span across the receptor. The first extracellular loop mutations reduce ligand affinity and the third intracellular loop mutations reduce signal transduction [100]. Recently, a cell-permeant small molecule that was a GnRH antagonist was shown to rescue most of the naturally occurring mutants by increasing their expression, presumably by stabilizing their intracellular processing and transport [101]. These small molecular ‘chaperones’ offer future therapeutic options for patients with GNRHR and other GPCR mutations.
GNRHR mutations can account for up to 40% of familial cases of nIHH and perhaps up to 17% of sporadic cases of nIHH [102]. Patients with GNRHR mutations present with a wide spectrum of reproductive symptoms ranging from severe hypogonadotropism including microphallus and undescended testes in males at birth to failure of pubertal development in adolescence [100] as well as infertility in adults [24,99,103]. However, partial defects are also seen with significant variations in phenotypes despite similar genetic functional defects [102]. This variability of clinical phenotypes presumably reflects several issues including the severity of intrinsic disruption of the GNRHR processing and/or function, the dosing of genes involved (heterozygous, biallelic/homozygous or compound heterozygous mutations), the coexistence of mutations in other GnRH deficiency causing genes (oligogenicity) [29], and as yet unapparent epigenetic and environmental factors.
TAC3 and TACR3
Using a similar strategy of studying consanguineous families with nIHH that led to the key discovery of the KISS1R mutations in human GnRH deficiency, Topaloglu et al.[104] recently reported loss-of-function mutations in both TACR3, the gene encoding a G protein-coupled receptor, neurokinin B (NKB) receptor and TAC3, the gene encoding NKB, the ligand for TACR3. This human genetic investigation highlights a hitherto unrecognized role of the TAC3/TACR3 pathway in regulation of the GnRH pulse generator. Neurokinin B belongs to a phylogenetically conserved family of proteins which also includes substance P, neurokinin A and hemokinin-1 [105]. TACR3 is predominantly expressed in the CNS including the hypothalamus [106] with high and preferential binding to NKB [107].
Male patients with TACR3 mutations characteristically have micropenis and fail to enter puberty during adolescence. These reproductive phenotypes strongly suggest an important role of the TAC3/TACR3 pathway in both the ‘mini-puberty’ of infancy and gonadotropin activation at puberty. None of the patients reported in the first human study had KS, suggesting a primary role of this TAC3/TACR3 pathway in functional integrity of the GnRH pulse generator. The receptor TACR3 is expressed on the GnRH axons and the KNDy (kisspeptin, neurokinin and dynorphin) neuronal population and neurons coexpressing KISS1 and TAC3 have been described in the arcuate nucleus and postulated to be the primary pathway mediating the sex-steroid feedback to the GnRH neurons [108,109]. However, murine deletion of the murine ortholog of TACR3 has not been associated with reproductive abnormalities as seen in humans [110]. Recently, Gianetti et al. have reported several TAC3/TACR3 mutations in a large outbred cohort of patients with nIHH [111]. In this detailed report, micropenis was observed in a majority of male subjects. In addition, evidence of neuroendocrine recovery of the hypothalamo-pituitary-gonadal axis was observed in a significant number of male and female subjects during adulthood. These observations strongly suggest that the TAC3/TACR3 signaling pathway is critical during the neonatal period and puberty, but is relatively less-critical in adulthood. No nonreproductive features have been reported in patients with TACR3 mutations, but TAC3 mutation have been associated with learning disabilities [104]. In addition, the potential oligogenic interactions of the TAC3/TACR3 pathway with the other genes related to GnRH ontogeny will need to be determined. The hypothalamic interplay between KISS1 and NKB is already well recognized [109,112,113] and this interaction is likely to be a subject of intense study in the coming years.
NELF
The human nasal embryonic LHRH factor gene, NELF, emerged as a strong candidate gene for KS following its strong association with axonal guidance of olfactory and GnRH neurons in mice [114]. Although a unique rare sequence variant in NELF has been reported in human GnRH deficiency, the biology of NELF in humans is still unclear [115]. However, the emerging theme of oligogenicity in human GnRH deficiency implies that NELF may be a critical modifier gene that synergizes with other pathogenic genes to result in the observed human phenotype [29].
Genetics of Other Multisystem Disorders Associated with KS/nIHH
Various multisystem disorders with overlapping features of KS/nIHH have been reported (table 1). Mutations of the DAX1 (NROB1) gene (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X chromosome gene 1), encoding a novel orphan nuclear receptor, have been identified in patients with X-linked adrenal hypoplasia congenita and nIHH [116]. Mutations in both leptin (LEP) and leptin receptor (LEPR) are associated with obesity and hypogonadotropic hypogonadism [117]. Heterozygous mutations in CHD7 (chromodomain helicase DNA-binding protein), a gene that encodes a large chromatin-modelling CHD7 protein, cause the CHARGE syndrome (eye Coloboma, Heart anomalies, choanal Atresia, growth and developmental Retardation, Genitourinary anomalies and Ear abnormalities) [118]. Some patients with CHARGE syndrome also share overlapping features of KS [119]. Recently, mutations in CHD7 have been identified in patients with both KS and nIHH [120], and in particular, KS/nIHH subjects who show presence of some of the CHARGE syndrome features such as deafness, dysmorphic ears and/or hypoplasia of the semicircular canals in addition to their hypogonadism, are more likely to harbor mutations in CHD7 [121].
Future Directions
In the last decade, the study of humans with GnRH deficiency has been prismatic in revealing the fascinating ontogeny of GnRH neurons. However, several key molecules/pathways that govern GnRH neuronal migration remain undiscovered and since pathogenic genetic mutations only account for approximately 40% of such patients, a substantial number of genes underlying GnRH neuron ontogeny are yet to be discovered. In addition, the emerging evidence of oligogenicity, modifier genes and potential gene-environment interactions emphasize the pressing need for an integrated investigatory approach encompassing detailed human phenotyping, use of next generation gene sequencing to aid novel gene discovery and the judicious use of bioinformatics. Although genotyping of GnRH deficiency is currently done predominantly in a research setting in most countries, the accumulating genetic knowledge will eventually allow the incorporation of genetic testing and genetic counseling into standard clinical care of these patients. The precise genetic basis of the onset of human puberty has been cited as one of the critical unanswered questions in the 21st century [122]. The prismatic human model of GnRH deficiency presents a unique biologic opportunity to unravel the molecular basis of the arousal of the GnRH pulse generator to signal puberty.



![Fig. 1. GnRH migration. a Migratory route of GnRH neurons in the embryonic mouse brain. GnRH neurons (black dots) originate in the medial wall of the olfactory placode, migrate along the olfactory axons and enter the brain perforating the cribriform plate. They then migrate to reach their final destination in the preoptic area (poa) of the hypothalamus. GnRH neurons on embryonic day 11 are seen in the vomeronasal organ (vno) and medial wall of the olfactory placode. In the 16-day-old fetal mouse brain, most of the GnRH neurons have reached the hypothalamus. gt = Ganglion terminale; ob = olfactory bulb. Reproduced with permission from Schwanzel-Fukuda and Pfaff [8]. b Cartoon of GnRH migration in humans. GnRH neurons originate in the embryonic olfactory placode and migrate along olfactory axons, penetrate the cribriform plate and migrate across the olfactory bulb, aided by various chemoattractive and repulsion factors and eventually reach the preoptic area of the hypothalamus, where they synchronize pulsatile GnRH secretion. Kisspeptin (KISS1) and the tachykinin (TAC) neurons serve as upstream modulators of GnRH secretion. Upon GnRH stimulation, the pituitary (PIT) secretes LH and follicle-stimulating hormone (FSH), which in turn regulate gonadal steroidogenesis and gametogenesis.](https://karger.silverchair-cdn.com/karger/content_public/journal/nen/92/2/10.1159_000314193/2/m_000314193_f01.jpeg?Expires=1716446195&Signature=kEFJQciIrsHFlQYEZFJCAt5ZOvz6Xp0se7xPtS2IL16JCQBKxV9fdHHezHIe5sT07mfQ9M6wj3SiReyPExfy3Qw8uYDYp0scv8geQvv9UY1-vZol0vPqsUsido2eU-jNgMZFLd62pikAqRdPVYbfU97CFGKQ3ZXmVVlWePj19sAwTnuiJSLn5cZpx4xms6a02xl~qzPWS8vJ48OqnTdDxEkQPt1hru8qEtkm~h9w0QCqZE4xqkt4MG6r2QrKMjI64ybK8IVdDwdFjDvqmZVpU6lemirfQ29fNUv3qNdiGECVcXpTX1Ia-vwv~qmJNETfC05V3wioJcsUs8ekUkJfJQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)





![Fig. 7. KISS1R and human GnRH deficiency. a Schematic of the predicted KISS1R protein displaying known mutations causing nIHH in humans. b Dose-response curves for the ligand-stimulated production of inositol phosphate in KISS1R mutant constructs, corrected for protein content. i Curve for the L148S mutation (three independent experiments, each performed in triplicate). ii Curve for the R331X mutation (two independent experiments, each performed in triplicate). iii Curve for the X399R polyA stop mutation (two independent experiments, each performed in triplicate). The percentages on the y-axis represent the percentages of the maximal stimulation for each GPR54 construct. iv The relative quantification of the wild-type and mutant GPR54 allele expression in lymphoblastoid cell lines as measured by quantitative RT-PCR. Reproduced with permission from Seminara et al. [50].](https://karger.silverchair-cdn.com/karger/content_public/journal/nen/92/2/10.1159_000314193/2/m_000314193_f07.jpeg?Expires=1716446195&Signature=LI5Q2b0TeO1Hs3sirgMNN3OJTB~0cYCARjdFMjN9qysi0RdxeJSemFNmsvCps5ofSch7CRvIXCEThW6eFN1XQ2EQPodmRQhuRTOYjAjkPWyOzhbny0JiuYMa0uDmYKWW9qzzOlM3YKyzVg1rjcT-Ic6UAL~vywIZ~XBT~vUprAq01Fs~lnWn4OvMKpsFqbJPsTc5WO6qFCm35xG0MourVKo~9EstVrv7YmShWuXQxicoGdg7R05bHcdtJ2HP319Ni5ecSR0GxDI1XNcF7cQaS48tqMLN6UwBP3dKrHoALVhmIS~nLOI164~eAos3-bk0z1H-w0vre--AzIJkDaRKOQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 8. FGF pathway in human GnRH deficiency. a Schematic of FGFR1 protein displaying mutations causing KS (blue) and nIHH (black). The extracellular domain of FGFR1 contains a signal peptide (SP), three extracellular immunoglobulin-like (D1, D2, and D3) domains, followed by the TM, and an intracellular domain comprising two tyrosine kinase subdomains (PTK). The acidic box (AB) and the docking protein FRS2 domain in the extracellular domain and intracellular domain, respectively, are specific features of FGF receptors. b Genomic structure and differential splicing of the human FGF8 gene. i Structure of the FGF8 gene. Boxes denote exons; lines denote introns. ii Schematic of the four FGF8 isoforms identified in humans with mutations identified to date are indicated by arrows and numbered according to the FGF8f and FGF8b protein isoforms (* homozygous mutations). Reproduced from Falardeau et al. [71].](https://karger.silverchair-cdn.com/karger/content_public/journal/nen/92/2/10.1159_000314193/2/m_000314193_f08.jpeg?Expires=1716446195&Signature=W1-~CwkYaNS9vp2TfxpBpR5QvbJUtXS7cHdoxXVeD~KdWDhxI0IA1zz4-OfKvaEKkjq51ZFjkvM7x4mOfMRQV9PF45NqgKS~ybdfhwBfVO1JR95~HtDfv8fwXPH7s3NooECR6Gy3GHnRr~UnQ-72wBVZu7BmXefTCLMA2uxFqNiD2nj3qk54elHkumBxQFuIi9XBnhQ9vT9hADlwrhkDYKc264qumFjBnPIHbkzd-ic1ET4gWzpYdf2oB-3d0wn1V-56-WRJaxgh4PLxYKCobFeVxjrWoq7Y5rAYcRz-nF~g44eRzwvpCwFKu1Ko~SM2y95IDw3sbuNpP2lq1eE1Lg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 9. Prokineticin 2 pathway and human GnRH deficiency. a Schematic of the predicted PROKR2 protein displaying mutations causing KS (yellow boxes) and nIHH (white boxes). b Morphological examination of wild-type PROK2+/+ and PROK2–/– mice showing hypoplasia of olfactory bulbs (red circle). Reproduced with permission from Ng et al. [79]. c In vitro activity of a selection of PROKR2 mutants in an intracellular Ca2+ assay. Reproduced with permission from Cole et al. [90].](https://karger.silverchair-cdn.com/karger/content_public/journal/nen/92/2/10.1159_000314193/2/m_000314193_f09.jpeg?Expires=1716446195&Signature=pyH7EBgbNWjkTzjN0zhYFO76Nb3WX1LyKlsnPHGPwL~zsjF8MwGmmQ52wX3fMPPv0Z4n3WXVy4fJTgEU2H-xAzor9-UFruzhFI0xns5b7pDWTSofUyKjyUG5eBE~3eMWAWXnfGdGeDldmrkpDMg059LV3Qmdo6MkaW91A2BPuJFApVnW6vg6Hlliz7uekTgonibp0KeNJFqLev6E5Fxp2cK3iM7yvcH6noAA3pZJRP76XRtBVVmXtxYJb6nvTBq1oAramWLttKGndCrUbeL-HMr13h0becVMyjbLbqSgKd4Bz-nPCZ7dg1qUtJ2jPuJDKOJevtcSAAABclS6RTp4kw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)