Abstract
Chlamydia trachomatis (CT) infections in women can result in tubal pathology (TP). Worldwide 10-15% of all couples are subfertile, meaning they did not get pregnant after 1 year. Part of the routine subfertility diagnostics is the Chlamydia Antibody Test (CAT) to decide for laparoscopy or not in order to diagnose TP. The CAT positive and negative predictive value is such that many unneeded laparoscopies are done and many TP cases are missed. Addition of host genetic markers related to infection susceptibility and severity could potentially improve the clinical management of couples who suffer from subfertility. In the present study, the potential translational and clinical value of adding diagnostic host genetic marker profiles on the basis of infection and inflammation to the current clinical management of subfertility was investigated. This review provides an overview of the current state of the art of host genetic markers in relation to CT infection, proposes a new clinical diagnostic approach, and investigates how the Learning-Adapting-Leveling model (LAL, a public health genomic (PHG) model) can be of value and provide insight to see whether these host genetic markers can be translated into public health. This review shows that the preliminary basis of adding host genetic marker profiles to the current diagnostic procedures of subfertility is present but has to be further developed before implementation into health care can be achieved. CT infection is an example in the field of PHG with potential diagnostic to be taken up in the future in the field of subfertility diagnosis with a time line for integration to be dependent on enhanced participation of many stakeholders in the field of PHG which could be advanced through the LAL model.
Chlamydia trachomatis Infection
Chlamydia trachomatis is the most common bacterial sexually transmitted infection throughout the world. An estimated 89 million cases per year worldwide are reported. The infection is often asymptomatic resulting in patients not seeking treatment. Untreated urogenital C. trachomatis may give rise to late complications, including pelvic inflammatory disease, ectopic pregnancy and tubal pathology [1,2,3,4]. The clinical course of chlamydial infections is heterogeneous i.e. transmission, symptoms, clearance, and development of late complications differ per patient [5,6,7].
Sexually acquired C. trachomatis is an important public health concern for its effects on reproduction. Women who develop late complications, such as pelvic inflammatory disease or tubal pathology, suffer considerable morbidity and emotional distress, and are a socio-economic burden [8].
To prevent such outcomes, early diagnosis is important. Currently, screening for tubal pathology is performed via laparoscopy, a procedure which is invasive and expensive, labour intensive and has a risk for surgical complications. This has resulted in extensive efforts to improve noninvasive diagnostic tests to decrease the risks of current screening methods.
Much research on bacterial components, clinical and environmental factors [9,10] has been done, but no definitive correlates of late complications have been identified [11]. For a variety of infectious diseases (e.g. malaria, hepatitis and meningococcal infections), it has been shown that host genetics play a crucial role in susceptibility to and severity of disease [12,13,14]. To estimate the role of genetics in course of infection, twin studies are a powerful tool, and Bailey et al. [15] have shown that host genetic factors contribute almost 40% to the variation in clinical course of Chlamydia infection. These results establish the potential importance of genetic studies. Den Hartog et al. [16] showed, in a cohort of subfertile women, that single nucleotide polymorphisms (SNPs) in several pattern recognition receptors (PRRs) increase the risk of developing tubal pathology following a C. trachomatis infection.
The innate immune response is the first line of defence against a C. trachomatis infection. PRRs recognize components of the bacterium, and SNPs in these genes may affect the functionality of these PRRs and may, therefore, increase the risk for development of late complications. In addition, SNPs in genes coding for cytokines involved in immune responses may also be influential. Recently, our group [17] reviewed the role of SNPs in PRRs and cytokines in relation to susceptibility to a C. trachomatis infection.
It is important that these scientific findings are utilized in the clinic and incorporated in public health policy. The Learning-Adapting-Leveling (LAL) model [18] is a model of translating scientific data from the lab through the market and implementing it into public health policy; it will be discussed later in this review as a possible way to assimilate new findings in clinical settings.
Outline of the Article
The overarching aim of the current review is to determine if C. trachomatis is one of the proof of principles in the field of public health genomics (PHG) with the potential to be taken up in the future in the field of subfertility diagnosis. Therefore, we provide an overview of the current state of the art on host genetic markers in relation to infection, propose a new clinical diagnostic approach in subfertility diagnostics based on this overview and describe how the LAL model (an integrated PHG model) can be of value to see if these host genetic markers can be translated from the lab to the market and implemented into public health.
Overview of CT Host Genetic Determinants of Infection
Several studies have shown the importance of host genetic variation on the clinical course of Chlamydia infections. This section will highlight recent findings, similar to a recent review [17], divided into detection of the pathogen Chlamydia by PRRs and the subsequent intercellular signalling by cytokines, with a focus on the innate immune system. Combined carriage of SNPs in so-called traits may exhibit a stronger influence on the course of Chlamydia infections, e.g. a reduced pathogen recognition capacity in multiple PRRs may result in higher susceptibility compared to the susceptibility when only one PRR has a reduced recognition capacity. Results for trait analyses are highlighted at the end of this section.
Pattern Recognition Receptors
Toll-Like Receptors (TLR)
TLRs are a much investigated group of receptors. Studies have shown that TLRs are essential in the host immune system by recognizing pathogenic components (pathogen associated molecular patterns (PAMPs) and danger associated molecular patterns (DAMPs) and inducing an immune response. These receptors are present on antigen presenting cells (APC) and epithelial cells; they reside both on the cell membrane and within cells. The TLR family has been studied in relation to various infectious and autoimmune diseases with varying associations [17,19,20]. TLRs 2, 4 and 9 are well-researched TLR family members. TLR2 and TLR4, both transmembrane pathogen receptors, recognize chlamydial peptidoglycan (PGN) and lipopolysaccharide (LPS), respectively. TLR9 is an intracellular receptor, recognizing CpG islands in bacteria.
Karimi et al. [19] investigated the role of 2 SNPs in TLR2 in C. trachomatis infected women and control groups: TLR2-16934T>A (rs4696480) and TLR2 +2477G>A (rs5743708). They did not find any association in TLR2 genotype distribution for both susceptibility to and severity of the infection. However, in haplotype analysis, they showed that haplotype TG was protective for developing tubal pathology. Laisk et al. [21] also evaluated the role of TLR2 +2477G>A (rs5743708) in developing tubal pathology and found no association, confirming the results of the single SNP analyses of Karimi et al. [19].
Den Hartog et al. [22] studied TLR4 +896A>G (rs4986790). The genotype distribution of this SNP in subfertile women with or without C. trachomatis infection did not differ. However, women with this SNP and who were positive for C. trachomatis IgG and cHSP60 IgG had tubal pathology. Results of this study are highly specific, but have low sensitivity, since not all women with tubal pathology had this combination of C. trachomatis and cHSP60 serology, and TLR4 +896 mutation carriage. Laisk et al. [21] also investigated the role of this SNP in developing tubal pathology; they did not find any association. Taylor et al. [23] studied a different TLR4 polymorphism, rs1927911, and found that the mutant genotype increases the susceptibility to C. trachomatis infections. Similarly, they found that the TLR1 rs5743618 TT genotype increased susceptibility to Chlamydia infections [23].
Ouburg et al. [24] studied TLR9 SNPs in a murine model, in a cohort of Dutch Caucasian women visiting a STD outpatient clinic and a cohort of subfertile Dutch Caucasian women. The overall genotype distribution did not differ between groups. However, haplotype analyses showed, though not statistically significant, that distribution of TLR9 haplotype -1486 T (rs187084), -1237 C (rs5743836), +1174 G (rs352139), and +2848 A (rs352140) was more frequently found in women who developed tubal pathology.
C-C Chemokine Receptor Type 5 (CCR5)
CCR5 is a chemokine receptor present on several immune cells, including monocytes, dendritic cells, microglial cells, T helper 1 cells, and macrophages. A 32-bp deletion within the CCR5 gene, CCR5 Δ32, results in premature termination of the protein, altering its function [25]. Barr et al. [26] demonstrated that this mutation has a protective effect against developing tubal pathology when both alleles are mutated. However, these results were not confirmed in a recent study [21]. For this inconsistency, the authors suggest that the ligand of CCR5, RANTES (CCL5), binds the CCR1 chemokine receptor as well, therefore inducing a normal response. Since they also used a different study population, they addressed the importance of group selection, and as a result this may be a reason for finding different outcomes [21].
Mannose-Binding Lectin (MBL)
MBL is important in the innate immune response. It binds to various carbohydrate structures of a.o. bacteria and either directly kills the pathogen or promotes phagocytosis [27]. Studies have shown that MBL inhibits a C. trachomatis infection [28]. Laisk et al. [21] investigated the role of 6 polymorphisms in the MBL2 gene, coding for MBL. They found that a hyperproduction haplotype of MBL2, HYA/HYA, was a risk factor for tubal pathology independent of a C. trachomatis infection. They also found this association, only smaller, in C. trachomatis infected patients with tubal pathology. They suggest that hyperproduction of MBL may affect epithelial cells within the genitourinary tract, inducing tubal pathology [21]. The low-producing MBL2 genotypes are associated with tubal pathology and adverse outcome of in vitro fertilization (IVF) treatment [29].
Human Leukocyte Antigen (HLA)
HLA codes for the major histocompatibility complex and thus, has an important function in the immune system. The HLA system has been linked to a variety of infectious diseases and disease outcomes. In the literature, a link between HLA-DQA1*0102 and HLA-DQB1*0602 alleles, and Chlamydia induced tubal pathology has been described [30]].
Another studied receptor is the major histocompatibility complex class I chain-related A (MICA), present on a.o. natural killer cells. When its ligand binds, activating signals for natural killer cells increase. Allele MICA*008 had a high negative correlation with C. trachomatis IgG antibodies. In this study, IgG antibodies were associated with tubal pathology. This group hypothesized that MICA alleles might play an important role in the development of tubal pathology. However, in the infertile women of their study, they could not establish an association between MICA alleles and tubal pathology with or without C. trachomatis IgG antibodies [31].
Cytokines
A variety of cytokines have been associated with disease and disease outcomes. These cytokines have important immunoregulatory functions, and alterations in function may, therefore, influence immune responses. Several cytokines have been studied in women who developed tubal pathology. Some associations were found: IL10 -1082 A allele together with HLA-DQA1*0102 and HLA-DQB1*0602 alleles; these were associated with severe tubal pathology [30,32]. Both the TNF-α -308 A allele and the IL6 CC genotypes were found to be associated with tubal pathology: the former as a risk factor and the latter as a protective factor for tubal pathology [32].
A statistically significant association between the IFNg+874 polymorphism and chlamydial tubal pathology was not found [32], nor for IL1B+3954, IL1B-511, and IL1RN gene polymorphisms [33]. However, a study performed in an ex vivo model showed that IL1, in the absence of its antagonist IL1RA, causes destruction of the ciliated cells in the Fallopian tubes [34]. In addition, SNPs in NLRP3, associated with hypoproduction of IL1β, is involved in tubal pathology [35]. Due to these findings, one may hypothesize that SNPs influencing IL1 functionality may affect the development of tubal pathology and the rate of severity.
The mutant allele of the IL12B rs3212227 SNP is associated with increased susceptibility to tubal factor infertility and with a more severe progression of disease [36]. The same group also demonstrated that IL10 and IFNG genotypes affect the lympho-proliferative responses in Chlamydia infections [37].
Trait Analyses
Den Hartog et al. [16] investigated the role of 5 SNPs in 4 genes assumed to play a role in C. trachomatis infection. The investigated genes were TLR4, TLR9, CD14, and CARD15/NOD2. The risk for development of tubal pathology doubled if a patient had 2 or more SNPs within the studied genes, compared to 1 SNP. In addition, when investigating only 1 SNP in TLR4 or CD14 [38], no association with tubal pathology was found. Due to a small sample size, no statistical significance was observed in the trait analyses; a statistical trend, however, was observed.
Ohman et al. [37] found that the combined carriage of specific IL10 and IFNG genotypes has an additive effect on the risk for Chlamydia infection. Atik et al. [39] demonstrated that combinations of SNPs affect the adverse outcome of ocular Chlamydia infections. They showed that trachomatous trachiasis risk decreased 5 times with the combination of TNFA (-308A), LTA (252A), VCAM1 (-1594C), and SCYA 11 (23T) minor allele, and the combination of TNFA (-308A), IL9 (113M), IL1B (5′UTR-T), and VCAM1 (-1594C). However, trachiasis risk increased 13.5 times with the combination of TNFA (-308G), VDR (intron G), IL4R (50V), and ICAM1 (56M) minor allele. Although these results are from ocular infections, one might hypothesize that similar effects might be observed in urogenital infections.
Although compelling, these results have to be confirmed in other studies, and additional SNPs have to be added in order to define the SNP profiles that are associated with and would help predict a patient's predisposition to Chlamydia infections and tubal pathology. This requires large cohorts which can be obtained via large consortia, in which different disciplines contribute to the overall goal.
Consortium Approaches: EU Framework Programme EpiGenChlamydia
To perform large scale typing for the identification of genetic biomarkers, large and clinically well-defined cohorts are needed. Toward this end, a small consortium was founded in 2005 with Dutch, Belgian and American partners with expertise on clinical, epidemiological, bacterial, animal, immunological, and host immunogenetic studies to have an integrated approach to study C. trachomatis infections, especially the clear inter-individual differences in the clinical course of infection. This consortium was named the ICTI consortium [40,41]. Members of the ICTI consortium applied for and obtained funding from the European Union as a large international consortium consisting of 20 partners, the EpiGenChlamydia (EGC) consortium [42].
This Chlamydia consortium was funded by the EU Framework Programme 6 under the Coordination Actions in functional genomics research for a period of 2.5 years and provided its closure report in 2010. The aim of this consortium was to structure transnational research to such a degree that comparative genomics and genetic epidemiology on large numbers of unrelated individuals could be performed with future funding. This funding made it possible for 20 groups from Europe, Africa and the USA to participate (see http://www.EpigenChlamy-dia.eu for details). The overall goal of the EGC consortium was to accommodate the optimal environment to build and prepare a consortium to reliably determine the genetic predisposition to infection in both ocular and sexually transmitted C. trachomatis. This will allow the development of diagnostic tools that can determine an individual's predisposition to infection and the risk to develop late complications. Further, it was hoped that the knowledge generated through this effort would contribute to the understanding of the Chlamydia- host interaction, in order to allow the development of novel tools for the detection and treatment of and vaccine development for C. trachomatis infections.
The EGC consortium has provided the final reports to the EU including state-of-the-art reports on the epidemiology of both ocular and sexually transmitted C. trachomatis infections [4], bacterial typing [43,44], immunogenetics [17], SNP genotyping strategies, and sample validation. Two deliverables were of major importance for the future success in translation of immunogenetic markers and for obtaining new funding for a biobank, consisting of physical and virtual sample collections, and a data warehouse in which genotyping data together with clinical and demographical data is merged and accessible.
The partners working on ocular Chlamydia diseases coordinated by the London School of Hygiene and tropical medicine (David Mabey and Robin Bailey together with their Gambian partners) have already defined and secured 1,500 case-control pairs (total n = 3,000). The scientific coordinator of the EGC consortium (S. Morré) together with Dutch collaborators have collected more than 7,000 specimens which are at present in use, while 10,000 specimens are available for further studies.
Currently, part of the consortium has obtained new funding from the EU based on Small-to-Medium-Enterprise collaborations with universities. This EuroTransBio grant has as main goal to develop a diagnostic test on the basis of human genetics and C. trachomatis serology to better assess the presence of C. trachomatis-associated tubal damage in subfertile women. This consortium, ending in the beginning of 2015, is in progress of performing large scale analyses of human genetic variation to identify novel genetic markers that are able to stratify patients with tubal pathology. The identified SNPs in the PRR genes have already shown to be highly predictive for the development of tubal pathology. However, single SNPs do not provide a high enough predictive value for a diagnostic test. By combining multiple identified and novel SNPs in the PPR genes and genes in linked pathways, and exploiting them as susceptibility markers, a highly predictive test for tubal pathology-based subfertility can potentially be developed. This will be discussed further in the next section.
Improvement of Subfertility Diagnostics Based on Host Genetics
Subfertility poses an enormous burden on healthcare and society throughout the world. Worldwide, 15% of couples trying to conceive suffer from subfertility [45,46]. In women, one of the major causes of female subfertility is tubal pathology [45]. In tubal pathology, C. trachomatis is the single most common cause for infertility [45].
From all subfertility problems in women, tubal damage is a common cause of infertility. It includes tubal obstruction and pelvic adhesions resulting from infection, endometriosis and previous surgery. The current diagnostic procedure for diagnosing this condition can be performed by sonohysterography/hysterosalpingo contrast sonography, hysterosalpingography (HSG), fertiloscopy, falloposcopy, or laparoscopy and dye hydrotubation, often using detection of IgG antibodies against C. trachomatis as first screenings tool.
Despite their effectiveness, the above-mentioned methods are costly and invasive [47] and not suitable for screening. There are several test methods available to assess the risk of C. trachomatis-associated tubal pathology in subfertile women. The reference standard for diagnosing tubal pathology in subfertile women is laparoscopy. However, laparoscopy has several disadvantages. First, it is an invasive, expensive procedure (on average 3,000 Euros, including additional costs) and requires general anaesthesia. Furthermore, it holds a 1.5% risk of surgical complications (e.g. bleeding, infection).
Since it is widely recognized that a C. trachomatis infection is the single most common cause of tubal peritoneal damage (WHO task force on the prevention and management of infertility [48]), detecting evidence of infection using serology is noninvasive, simple and quick to perform [47]. As such, Chlamydia serology is often used as a first screening test for tubal damage in infertile women but has a limited sensitivity of 50-60%.
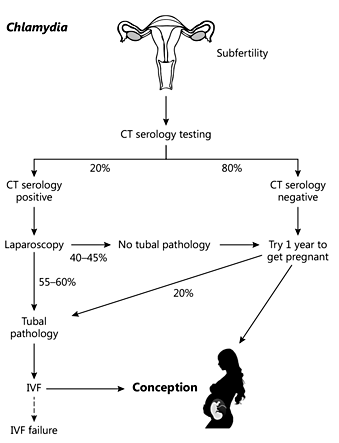
Currently, women with subfertility are screened for a C. trachomatis using serology (see fig. 1). Serologic testing (CAT: Chlamydia antibody testing) for C. trachomatis is based on micro immunofluorescence assays. Elevated titres of IgG are highly predictive for infection with C. trachomatis. These serologic assays focus mainly on the major outer membrane protein A, which is an antigen present in the outer membrane of a chlamydial particle. When the serology outcome is negative, no further action is taken, and the couple is asked to try for one more year to get pregnant. Some women undergo HSG. If the outcome is negative (in most of the cases), they try as well to conceive for one more year. If positive (in around 5-7% of the cases) a laparoscopy will be performed, and up to 5% will have tubal pathology and will proceed to IVF procedures. Since HSG does not identify many new cases (5% of HSG positive cases), its positive predictive value is almost identical to serology. The low specificity of the test also causes misdiagnosis because women that are negative in the serology test may in fact have tubal pathology in up to 20% of cases (percentages based on the cohort described in ref [16], personal communication).
The relation between serology testing (Chlamydia antibody testing) and laparoscopy and the percentages of misdiagnosis. CT = C.trachomatis; IVF = in vitro fertilization.
The relation between serology testing (Chlamydia antibody testing) and laparoscopy and the percentages of misdiagnosis. CT = C.trachomatis; IVF = in vitro fertilization.
If the serology outcome is positive, laparoscopy will be used for further diagnosis. When tubal damage is detected using laparoscopy, it is likely that an IVF procedure will be initiated to get pregnant. One of the drawbacks of serology is that it comes with limited sensitivity: 55-60% of the Chlamydia serology positive women actually have tubal pathology. Subsequently, this means that 40-45% of the women are serology positive and undergo laparoscopy while there is no tubal disease. In conclusion, in the serology negative and positive groups, a significant percentage of the women get either unneeded laparoscopies or get misdiagnosed.
Given the prevalence of subfertility, each year an estimated number of 300,000 women visit the general practice with C. trachomatis-associated infertility in Europe. Using the current diagnostic procedures, there is still a substantial subset of women that are misdiagnosed. This poses an enormous psychological burden on these women. Additionally, there are tremendous economic costs associated with the disease.
Despite the rapid development in the field of human reproductive medicine, there is still a medical need for diagnostic tools that are able to stratify clinically relevant C. trachomatis infections. The diagnostic test envisioned should be able to predict C. trachomatis-based tubal pathology and subsequent infertility by complementing serology in such a way that not only the sensitivity, but also the positive and negative predictive values increase significantly.
Epidemiological studies have demonstrated a 40% inheritable component for Chlamydia infections in humans [15]. This suggests the need for studies to identify which genes are responsible for this 40% component of risk. The innate immune system plays a pivotal role in the first recognition of Chlamydia and the subsequent immune response.
Since a panel of human PRRs are involved in the recognition of Chlamydia, we hypothesized and have shown that carrier traits (i.e. carrying multiple SNPs in multiple genes) result in a higher aberrant immune response as compared to single gene association studies. Subsequently, these traits synergistically increase risk for tubal pathology following C. trachomatis infection. Of 227 subfertile Dutch women, we performed genotyping of PRR genes TLR9, TLR4, CD14, and CARD15/NOD2 looking for common versus rare alleles [16]. Subfertility was defined based on laparoscopic grade of tubal pathology including extensive peri-adnexal adhesions and/or distal occlusion of at least one tube. Being a carrier of several rare alleles was more frequent in women with tubal pathology (who had elevated serum IgG titers against C. trachomatis). We showed that after C. trachomatis infection (these infections defined as CT IgG titers >32), subfertile women carrying >2 SNPs in PRR genes were at increased risk for tubal pathology compared to women carrying <2 SNPs (73 vs. 33% risk). This association was not found among women without IgG antibodies to C. trachomatis. Thus, adequate recognition of C. trachomatis by receptors in the genital tract is an important step in the immune response and may play a role in protecting the host against developing late sequelae following infection.
The single SNP frequencies of TLR2 SNPs did not alter the risk for tubal pathology in subfertile women, but, combined into specific haplotypes, carriage of haplotype I significantly reduced the risk of developing tubal pathology after a C. trachomatis infection (p = 0.015, OR = 0.28). This haplotype also showed a significant trend in an inverse association with disease severity (asymptomatic > symptomatic > tubal pathology; ptrend = 0.021) [19]. In addition, PRRs, cytokines and chemokines play an essential role in the immunopathogenesis of C. trachomatis infections. The chemokine receptor, CCR5, is crucial for T cell activation and function, since its deficiency causes suppression of T cell responses. We showed that among patients with anti-chlamydial IgG responses, tubal pathology correlated with a low incidence of the CCR5Δ32 deletion (7%), while women without tubal pathology had a higher incidence of the CCR5Δ32 deletion (31%), as compared to controls (19%) [26]. Thus, inflammation associated with CCR5 may predispose to development of complications of C. trachomatis infection. Recent findings show a similar pattern for CXCR5 [49].
Therefore, single SNPs, haplotypes, and eventually larger genetic traits based on genetic variation in multiple genes can potentially be used as susceptibility or severity markers for tubal pathology as a result of a C. trachomatis infection. By combining multiple SNPs in one diagnostic test, high predictive values can be achieved which should be suitable as a future companion diagnostic in the diagnosis and treatment strategy for subfertility.
PHG Approached and Translation into Public Health
Findings in the field of immunogenetics of C. trachomatis infections are of high relevance for public health and healthcare in general. Such results contribute to the understanding of infection with this agent, which is worldwide the leading cause of preventable blindness and the most prevalent sexually transmitted disease that is strongly associated with ectopic pregnancy, tubal infertility and pelvic inflammation. Furthermore, our findings provide new insights into the pathways that help explain individual heterogeneity in the clinical course of C. trachomatis infection and the possible development of more targeted and personalized approaches in the prevention, diagnosis and treatment of disease.
To improve the health outcomes associated with C. trachomatis infections, there is an urgent need to timely translate immunogenetic findings into the healthcare system. The specialty having this task is called PHG, which is defined as ‘the responsible and effective translation of genome-based knowledge and technologies into public policy and health services for the benefit of population health' [50]. In the context of C. trachomatis infections, such a public health initiative has not been advanced, despite sufficient data and the significant need to do so. However, significant public health strategies need to be implemented very early after the discovery phase, and proofs of concept need to be obtained to promote a faster translational process not only from bench to bedside, but also from bedside to healthcare. Possible bottlenecks for implementation need to be identified.
Developing diagnostic tools based on host genetic predisposition can help determine an individual's risk (as well as late complications) of infection. However, moving forward with such a plan has hurdles along the path. We have seen that C. trachomatis infections, symptoms and complications can differ between individuals on the basis of host genetic factors, ethnicity and environmental factors. And based on this review and on the literature, it seems likely that diagnostic assessment will allow for inclusion of a large number of case-specific variables (i.e. more stratified) that may even become ‘truly' personalized in the near future with the incorporation information derived from dynamic fields of investigation such as systems biomedicine and epigenomics [51]. The major obstacle to implementation is not the CE (Conformtie Europeene) IVD or FDA approval of diagnostic application, but originates with the healthcare integration [52] and policy embedment processes.
In general, a timely translation with direct implementation by the healthcare systems is low [53], which is illustrated by the large amount of data present in the literature [54], patents [55,56] and marketed products [57,58] compared to technologies being used in hospitals [53,59]. In order to minimize failure, it is important that researchers take into account the policy aspect and the acceptance of diagnostic applications in the healthcare systems [52] itself, instead of just considering the market.
In order to move into the healthcare systems, it is important to think from the decision-making and policy implementation perspective. Health policy is generally developed through evidence-based interventions and around general public health instruments, such as Health Technology Assessment (HTA) among others [60,61]. We use the term Public Health Assessment Tools (PHAT) [52] when we refer collectively to Health Needs Assessment, HTA and Health Impact Assessment. Health Needs Assessment is a systematic method of reviewing the health issues facing a population, leading to agreed upon priorities and resource allocation that will improve health and reduce inequalities [61]. HTA is a multi-disciplinary field of assessment that evaluates the medical, economic, social, legal, ethical, and other implications of the incremental value, diffusion and use of a technology in healthcare [62]. Health Impact Assessment is a combination of procedures, methods and tools by which a policy, programme or project may be judged as to its potential effects on the health of a population and the distribution of those effects within the population. For simplicity, we will utilize HTA [63]. This assessment is generally done by HTA professionals in the form of systematic reviews of an emerging or new technology/tool with recommendations forwarded to decision-makers. Based on these recommendations, a technique, technology, tool, or process is implemented in the policy of healthcare, and these policy decisions determine the acceptability, the applicability in healthcare and the reimbursements. This in our observations has been generally neglected [52] by the academic-industrial complex [64]. Therefore, it is of uttermost importance to take into consideration the HTA process while developing a tool, diagnostic kit or technology, thus, in order to efficiently, effectively and in a timely manner introduce an innovation into the healthcare system. Although HTA or PHAT themselves are not sufficient when looking at the whole translational pipeline from bench to healthcare, HTA needs to be streamlined and integrated with the technology transfer process, which is the process of translating an idea into an innovative product on the market.
In the case of C. trachomatis, it can be said with some certainty that HLA, specific cytokines including IL-10, and several TLRs play a role in infection and disease progression. These factors will likely vary between ethnical groups. For example, the most important TLR4 SNP, associated with tubal factor infertility in Caucasians, does not exist in Chinese people at all. Furthermore, the current needs of the population, relevant technologies on the market and prioritizing technologies based on applicability, the target audience, and stakeholder involvement including patient groups need to be taken into consideration. Also, the analytical validity (the ability of a test to accurately and reliably measure the genotype of interest) [65], clinical validity (the ability of the test to detect or predict the phenotype of interest) [65] and clinical utility (the likelihood that the test will lead to an improved outcome and incorporates assessment of the risk and benefits of genetic testing as well as economic evaluation) [65] of the diagnostic tool will have to be considered. In addition, by performing HTA, the economic, ethical, legal, and social implications will have to be thoroughly addressed in order to preempt any HTA related assessments by health policy-makers for technology integration. This collectively covers many aspects of HTA investigation.
Healthcare aims to improve health of the populations and that the perspective from which the healthcare decision-maker works is based on HTA recommendations. Therefore, at the end of the day, the approval of the technology lies in its clinical utility, since the analytical validity and clinical validity have been more or less addressed. Furthermore, equal weight is given to its ethical, economic, legal, and social implications (ELSI) in society. Generally, ethical, legal and economic issues are dealt with during the development of a diagnostic tool or technology; however, the social implications may be overlooked. This can be the downfall of the tool in healthcare implementation. Also, ELSI can be limited to one perspective: the industrial and not the population based perspective. Taking into account future developments such as ‘truly' personalized medicine moving from clinical utility to personal utility and the use of ‘personal-genome tests' [66], current HTA as a tool for decision-making will be challenged [67]. Current HTA evaluates a technology on the population or subpopulation, but not on an individual level, which will be the need in the era of ‘truly' personalized medicine.
When talking in terms of economics, reimbursement through insurance companies becomes important, which is sometimes addressed during the development of a technology. Financially stable patients with no need of reimbursement, early adopters or ‘trendsetters' do not guarantee acceptance by decision-makers as this does not represent the majority. HTA analysis is done based on the needs of the population as a whole and not based on a few elite. Therefore, these ethical questions have to be addressed here as well. HTA professionals generally prioritize technologies listed by them and investigate one or a few of them based on what they deem relevant to the current population need, which also has to be taken into account during the development of the diagnostic tool. This can be done via comparison to what already exists on how one can get a competitive advantage based on the current population need and through consultation with patient groups. This brings in another important aspect: the involvement of stakeholders. These stakeholders represent the needs of the population and should be given preference through the development of the diagnostic tool or technology. All these several aspects should be taken into account while developing a specific diagnostic kit for C. trachomatis subfertility, in order to decrease the chance of failure, if not guarantee its success in healthcare implementation. As a result among other factors, there is generally a delay in uptake and wide usage of diagnostic applications and technologies in healthcare systems and hospitals. Consequentially, by the time the technology or kit reaches the healthcare system, it becomes inferior in terms of efficiency and effectiveness, given the exponential growth of newer versions of emerging technologies on the market. The latter again goes through this delaying process.
Recently, a new framework or model [52] has been developed which addresses all the above mentioned issues and others not mentioned here with regard to healthcare implementation. The LAL model brings together for the first time 2 separate entities, namely technology transfer and PHAT in a pseudo-parallel initiation. Through this, the model promotes early on involvement of all stakeholders (including academics, industry, patient groups, insurance, policy makers, doctors, HTA professionals, etc.) via public-private partnership, consultation, bilateral communication, exchange of information, feedback loops, and relevant lobbying. The model ensures that through the technology transfer pipeline, all PHAT aspects are addressed, and it also encompasses the Public Health Genomics Wheel [68] as a reference frame which demonstrates the essential tasks of Public Health for integration of genome-based technologies ensuring all possible gaps are addressed. This ensures that by the time the technology or diagnostic application is developed, it conforms to the standards required by decision-makers based on population needs. This decreases the likelihood of rejection as a relevant tool for healthcare. The model also puts emphasis on the value of information (how much a decision-maker is willing to pay to come to a decision) [69]. This is in terms of the end-user clarity, including ease of use, relevance to the patient and doctors, and any legal issues restricting use of the diagnostic application widely. As a consequence of the value of information, adaptations in this case to the diagnostic application can be made accordingly. Through this process, by the time the technology or diagnostic application is developed, it meets all conditions of healthcare policy, therefore, facilitating timely uptake. We believe this overarching LAL model can ease the bottleneck of a real-time uptake by hospitals as well as help industry to come to an early-on strategic decision on the new technology and thus, save on resources [52]. The LAL model seems an appropriate tool and framework for the development of the Chlamydia diagnostic kit that will ensure all issues of healthcare, as well as public health conformity and industrial requirements, are met and addressed. As a result, by the time the diagnostic kit is ready, it can be made immediately available for widespread use throughout the healthcare system. This can help industry to tap into a generally wider consumer market than traditionally accessed as well as help decrease the burden of disease that more would result from delay in the technology.
It becomes obvious, that public health approaches need to adjust to these developments. Thus, PHG in the future will be quite different from PHG in the past [51].
Rapid scientific advances in genomics and its application to epigenomics, microbiomics and systems biology not only contribute to the understanding of disease mechanisms, and to the characterization of each person's unique clinical, genomic, and environmental information, but also provide the option of new promising applications in patient and human health management during the whole life-course. In fact, what was just a decade ago a distant vision for a new era of public health, in which advances from the -omic sciences would be integrated into strategies aiming at benefiting population health, is now the soon-to-be realized development of effective personalized healthcare that will be ‘truly' personalized medicine. The utility of most genetic tests and biomarkers is still not evidence-based enough. In the personalized medicine setting, the traditional assessment and evaluation tools are inadequate. We clearly face the need for a new paradigm because as we start to understand, for example, that what we call common complex diseases might be a sum of ‘rare diseases'; we move from risk factors to individual pathways or networks, and from that perspective, we move from clinical utility to personal utility [70]. However, the real paradigm shift depends on the willingness to restructure policies and on the ability to train practitioners from various professions. P4 Medicine is a future vision defined by biologist Leroy Hood, and is short for ‘Predictive, Preventive, Personalized, and Participatory Medicine' [71]. The premise of P4 Medicine is that, over the next 20 years, medical practice will be revolutionized by biotechnology, to manage a person's health, instead of manage a patient's disease. Although probably not around the next corner, there is a clear urgency to prepare healthcare systems and policy-makers in advance of the inevitable.
The implementation of PHG requires increased concerted actions not only on the global (http://www.graphint.org), but also on the European level. The Public Health Genomics European Network (PHGEN I), which is funded by the General Directorate for Health and Consumer Protection (DG SANCO) (http://www.phgen.eu), initiated the National Task Forces on PHG in over 15 EU Member States. Due to these initiatives, the National Institutes of Public Health took a leading role in PHG in Finland, Belgium, the Netherlands, Croatia, Poland, or Germany. PHGEN II has developed ‘European Best Practice Guidelines for Quality Assurance, Provision and Use of Genome-based Information and Technologies' [72], which will assist the European Member States. The LAL model has also been integrated in these guidelines.
A major international research consortium called ‘Information and Communication Technology for Future Medicine (ITFoM)' anticipates the medicine of the future, based on molecular, physiological, anatomical and environmental data from individual patients (http://www.itfom.eu). The ‘ITFoM project' will create the entirely new ICT that will make it possible to make general models of human pathways, tissues, diseases, and ultimately of the human as a whole. Patient-individualized versions of ICT replica (‘virtual patients') will be used to identify personalized prevention and therapy schedules and side effects of drugs [73]. The LAL model will play an important role in this project to make sure that by the time the technology is rolled out, it is adapted and conformed for real-time integration in healthcare.
Conclusions and Future Prospects
We investigated the potential translational and clinical value of adding diagnostic host genetic marker profiles relating to infection and inflammation to the current clinical diagnosis and management of subfertility which is based on serology and laparoscopy. It is clear from a large body of evidence that host genetic factors play a role in the susceptibility to and severity of C. trachomatis infection, as shown by twin studies and many candidate gene studies. To bring this current host genetic work to the next level, large scale SNP typing and SNP identification in confirmation cohorts is essential and is in progress. This work will provide insight into what type of host genetic profile can help improve subfertility diagnoses and whether the added sensitivity, specificity, and positive and negative predictive value will realize the hope that carrier traits will significantly increase the ability to predict and identify those at greatest risk of severe complications from C. trachomatis infection. In addition, as shown by the LAL model, stakeholders have to be informed and participate early-on in the potential implementation of these findings, a major task and challenge for the field of public health genomics which can be realized through the LAL model.
Acknowledgement
The authors thank the Public Health Genomics European Network (PHGEN) for commissioning work on assessment instruments in Public Health Genomics within the European Union. This work was supported by two grants of the European Commission (DG SANCO, Agreement PHGEN I 2005313 and Agreement PHGEN II 20081302). The aim of this work is in line with the European EpiGenChlamydia Consortium which was supported by the European Commission within the Sixth Framework Program through contract LSHG-CT-2007-037637. See http://www.EpiGenChlamydia.eu for more details about this consortium.
References
J.A. Lal and J. Malogajski contributed equally to this article.