Abstract
Nutritional research is undergoing a remarkable transformation driven by new technological tools. Because of the complexity of the components present in food and how they may interact with the biochemical networks of living organisms, nutrition cannot be considered a reductionist discipline. More holistic approaches, which are capable of gathering comprehensive, high-throughput amounts of data, appear to best enhance our understanding of the role of food in health and disease. In this context, global metabolite analysis, or ‘metabolomics', is becoming an appealing research tool for nutrigenomics and nutrigenetics scientists. The purpose of the present review is to highlight some potential applications of metabolomics in nutrition research.
Metabolomics, Metabolome, and Metabolic Phenotypes
Metabolomics can be defined as the screening of small-molecule metabolites present in samples of biological origins (plants, animals, or microorganisms) (fig. 1) [1,2,3]. The characterization of all the metabolites (‘metabolome') (fig. 2, 3) can provide a snapshot of the metabolism and a molecular fingerprint. Such a characterization is, consequently, an index, or biomarker, of a biological state of an organism [1,2]. By comparing metabolome profiles (metabolic phenotypes or ‘metabotypes'), we can determine patterns of variations between different groups: healthy versus diseased, control versus treated, wild-type versus genetically modified [1,2,3]. In addition, metabolomics can be used to monitor the outcome of treatment strategies, such as pharmacological or dietary interventions, by observing whether the metabolic phenotypes of treated, diseased patients shifts in the cluster of healthy subjects [1].
The growing field of metabolomics. The advances in metabolite analysis led to the emerging field of metabolomics. An up-to-date search using PubMed shows an exponential growth in the number of publications containing the term ‘metabolomics' OR ‘metabolomics' OR ‘metabonomics'.
The growing field of metabolomics. The advances in metabolite analysis led to the emerging field of metabolomics. An up-to-date search using PubMed shows an exponential growth in the number of publications containing the term ‘metabolomics' OR ‘metabolomics' OR ‘metabonomics'.
Metabolites. Metabolomics aims to screen metabolites in biological samples. Metabolites can be divided into water-soluble, or hydrophilic, compounds (e.g., nucleotides, amino acids, and carbohydrates; a) and water-insoluble, or hydrophobic, compounds (e.g., mostly lipids; b). Fatty acids are the building blocks of most lipid structures. Saturated fatty acids do not contain double bonds, while monounsaturated fatty acids and PUFAs contain one or more cis double bonds. There are two types of PUFAs: omega-6 and omega-3, named according to the position of the first double bond in the carbon chain, starting from the methyl end of the molecule. Each PUFA family contains both short- and long-chain PUFAs (Box 2). Other dietary components such as terpenes (e.g., carotenoids such as lycopene) and vitamin E (e.g., alpha-tocopherol) are mostly insoluble in water and classified as lipids.
Metabolites. Metabolomics aims to screen metabolites in biological samples. Metabolites can be divided into water-soluble, or hydrophilic, compounds (e.g., nucleotides, amino acids, and carbohydrates; a) and water-insoluble, or hydrophobic, compounds (e.g., mostly lipids; b). Fatty acids are the building blocks of most lipid structures. Saturated fatty acids do not contain double bonds, while monounsaturated fatty acids and PUFAs contain one or more cis double bonds. There are two types of PUFAs: omega-6 and omega-3, named according to the position of the first double bond in the carbon chain, starting from the methyl end of the molecule. Each PUFA family contains both short- and long-chain PUFAs (Box 2). Other dietary components such as terpenes (e.g., carotenoids such as lycopene) and vitamin E (e.g., alpha-tocopherol) are mostly insoluble in water and classified as lipids.
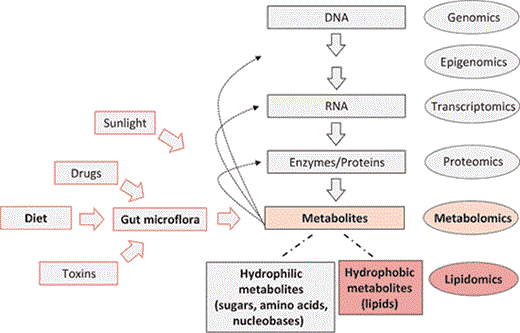
Metabolomics in systems biology. Genes (DNA) encode mRNAs that, in turn, encode proteins that collectively, and together with environmental factors (e.g., diet), lead to the metabolite inventory of a cell, tissue, or body fluid. Metabolites, in turn, can regulate gene expression, enzymatic activities, and protein functions. Among the metabolites are lipids. Novel approaches now allow for qualitative and quantitative measurements at each level on global scales (genomics, epigenomics, proteomics, and metabolomics). Lipidomics can be viewed as a subdiscipline of metabolomics under the umbrella of systems biology.
Metabolomics in systems biology. Genes (DNA) encode mRNAs that, in turn, encode proteins that collectively, and together with environmental factors (e.g., diet), lead to the metabolite inventory of a cell, tissue, or body fluid. Metabolites, in turn, can regulate gene expression, enzymatic activities, and protein functions. Among the metabolites are lipids. Novel approaches now allow for qualitative and quantitative measurements at each level on global scales (genomics, epigenomics, proteomics, and metabolomics). Lipidomics can be viewed as a subdiscipline of metabolomics under the umbrella of systems biology.
Unlike the genome, which remains static, the metabolome reflects both genetic and environmental components, including drugs, contaminants, gut microflora activity, and, most notably, diet (fig. 3) [4,5]. Thus, comprehensive metabolite profiles can offer a level of description of a biological system that transcends pure genetic information and more closely reflects the ultimate phenotypes. Together with other ‘-omics' disciplines such as genomics, transcriptomics, and proteomics, metabolomics is becoming an integral part of a systems biological approach for investigating organisms (fig. 3) [3,6,7].
Metabolomics in Nutrition Research
For millennia, besides providing a source of energy, food has been known to modulate health and well-being. The ancient eastern civilizations of Egypt, Persia, India, and China used food as medicine to treat and prevent disease. The father of Western medicine, Hippocrates, underscored this conception of food as evidenced by his renowned statement, ‘Let food be thy medicine and medicine be thy food.' It is still mostly unclear, however, which components in food - and through which mechanism of action - affect health and disease.
Historically, nutrition research scientists mostly used a reductionist approach, focusing on analyzing only a handful of food metabolites. A more holistic approach, however, seems better suited to understanding the interactions that can occur between the variety of chemical components found in the food and the biochemical networks of complex organisms. As we move into a new era of technology, the potential to accurately and rapidly measure hundreds of individual molecular species provides novel opportunities for nutrition research [8,9,10,11].
Metabolomics tools are now being applied to the analysis of food components, the identification of their metabolites in body fluids and biological tissues, the evaluation of their bioavailability and metabolism, the role of gut microflora, and the physiological response to a particular diet regimen, food, or nutraceutical (Box 1) [12,13,14,15,16,17]. Many reports referenced throughout this review provide the proof-of-principle that metabolomics is set to be a key tool for nutrition research.
Box 1. Nutraceuticals

Food Metabolome
Our limited understanding of the effect of nutrition on our well-being and health arises in large part from our limited knowledge about the molecular composition of food. With the advent of metabolomics, foods and beverages are now being analyzed with considerably more chemical detail, and thousands of chemical entities are being detected or identified in certain foods. The collection of all food components, both natural and non-natural, is often referred to as the food metabolome. The ultimate knowledge of the food metabolomes could provide a piece of critical information for the study of complex interactions between nutrition and health [12,13,14,15,16].
Although the applications of metabolomics in food and nutrition research are still in their infancy, they have high potential as shown in some examples reported below.
Natural Food Components
Until just recently, the analysis of food was limited to estimate its nutritional value based on the content of six broad categories: carbohydrates, fats, proteins, water, vitamins, and minerals.
Metabolomics is transforming modern nutrition research by looking beyond the plain list of nutrients, the number of calories, or the ratio of proteins to carbohydrates or fats. Indeed, evidence is growing that within the broad classes of nutrients, subclasses of metabolites could have opposite effects on human health (Box 2).
Box 2. The nutrition facts on fat

Furthermore, metabolomics is helping to explore the thousands of non-nutrient components that, although not required by the human body for sustaining life, could affect human well-being and health [18]. A large proportion of the food metabolome, indeed, consists of phytochemicals. Some of the well-known phytochemicals, such as the lycopene in tomatoes, isoflavones in soy, and polyphenols in fruits, are responsible not only for the organoleptic properties like flavors and aromas of the plants, but also for their health properties [18,19]. Notably, phytochemicals are rarely absorbed and excreted in their ingested forms, but extensively metabolized in the body. Therefore, only a limited portion of the theoretical 200,000 structures of phytochemicals have been characterized, suggesting that many remain for discovering [18].
Non-Natural Food Components
Metabolomic screenings are able to find a large list of environmental chemicals such as herbicides, insecticides, antimicrobials, fungicides, and toxins in foods and beverages [20,21,22,23,24]. The screenings also reveal contaminants that might affect health. These contaminants are derived from consumer products like face creams, soaps, disinfectants, flame retardants, antibiotics, fertilizers, drugs, and thousands of approved food additives and preservatives, whose effect on human health and environment remain mostly unknown [20,21,22,23,24,25].
Databases
A growing number of natural and non-natural food components and the products of their biotransformation are being annotated in publicly accessible databases [26]. When such databases are queried for a monoisotopic mass or elemental formula, they return a list of possible metabolites, with their chemical structures, biological properties, spectral data, related metabolic pathways, occurrence and concentrations in foods, and effects on health. Alternatively, when queried by food, such databases return a list of molecular components and their human metabolites likely to be present in biofluids after their consumption. Some example of current or upcoming food databases are provided below.
Recently, the US Department of Agriculture updated one of the major sources of food composition data in the United States, which is the National Nutrient Database for Standard Reference Composition of Foods Raw, Processed, Prepared. This database provides nutrient information on over 8,000 food items and up to 146 food components (http://ndb.nal.usda.gov/).
A similar comprehensive effort in Canada by Dr. David Wishart Research Group at the University of Alberta to annotate the ‘food metabolome' is under way (http://www.foodb.ca/). This food component database, called FooDB, will eventually provide information on over 28,000 food components and food additives, including many of the constituents that give foods their flavor, color, taste, texture, and aroma. Notably, the same group undertook a similar effort in 2005 to annotate the human metabolome, which, as of now, has catalogued more than 40,000 different chemicals found in the human body (i.e., Human Metabolome Database, HMDB; http://www.hmdb.ca/) [27].
Finally, various European research groups are currently working on databases tailored to the phytochemical components of the food metabolome. For example, Phenol Explorer contains values for 500 dietary polyphenols and their known human metabolites in over 400 foods [26,28]. This database has been developed at the INRA, Unité de Nutrition Humaine (Centre de Recherche de Clermont-Ferrand, France) in collaboration with the University of Barcelona under the supervision of Dr. Augustin Scalbert at the International Agency for Research on Cancer (IARC) in Lyon, France. A similar effort is under way as part of the Phenotyping using Metabolomics for Nutritional Epidemiology (PhenoMeNep) project funded by the French National Research Agency. This upcoming database, called PhytoHUB, will contain a comprehensive inventory of dietary phytochemicals and their human metabolites, using structures obtained from both previous publications and in silico predictions [29].
Engineering Food Metabolomes
The characterization and modulation of metabolic phenotypes helps the food industry to optimize organoleptic properties in food products and increase the abundance of healthful compounds in them. Some representative examples are provided below.
Metabolomics has the potential to inform the breeding selection of crops and unintended effects of genetic modification, representing an important addition to the tools currently employed in the genomics-assisted selection for crop improvement (European Food Safety Authority, EFSA, 2006) [3,20,30,31,32]. For example, metabolomics analyses of crops grown under different conditions highlighted that environmental factors, such as growing seasons or locations, can have a greater influence on the overall molecular composition than a difference in genotype [32].
In a different application, the characterization of metabolic phenotypes can be used to determine feeding optimization of livestock and agriculture [20,33]. For example, a recent report indicates a higher omega-6 versus omega-3 fatty acids ratio (Box 2) in farm raised compared to wild-caught fish [34]. Similarly, metabolomics approaches have been used to compare organic versus non-organic food [30,33]. For example, recent studies reported that organic cultivation was found to provide tomatoes with a significantly higher content of antioxidant phenolic compounds than conventional farming [33,35]. Finally, metabolomics allows scientists to discover and characterize the chemical modifications that result from food processing, which could significantly alter the molecular content and health properties of food [19,36,37]. The molecular composition of food products, indeed, can be significantly affected by different methods of food preparation (e.g., frying vs. baking, steaming vs. boiling) and preservation (freezing, drying, smoking, and refrigerating) [20,36,37]. Recently, a metabolomics approach was used to determine the molecular difference between wholemeal and refined pasta samples, finding that the wholemeal pasta was richer in many classes of compounds such as phytosterols, policosanols, unsaturated fatty acids, amino acids, carotenoids, and minerals [36]. Similar applications are starting to show the high potential of metabolomics for investigating the process-related food transformation [37].
Dietary Biomarkers
The characterization of food metabolomes is leading to the discovery of food-specific biomarkers, which are indicators of diet exposure and food consumption [38]. Already, metabolomic screenings revealed urinary markers associated with an individual's dietary intake of meat-rich diets (1-methylhistidine), vegetable-rich diets (phenylacetylglutamine), citrus (proline betaine), oily fish (methylhistidine), coffee (dihydrocaffeic acid derivatives), and tomato (phenolic metabolites) [38,39,40]. Similar quantitative screening of metabolites could soon facilitate the monitoring of food consumption in epidemiological or dietary intervention studies, supporting the use of diet questionnaires [15,39,41,42,43]. Furthermore, the development of rapid and inexpensive assays for biomarkers of food intake of health relevance could be used to routinely assess nutritional deficiencies and unbalances in large population cohorts [8].
Dietary Metabolites and Cellular Metabolism
Epidemiologic studies comparing countries reporting low and high incidences for particular diseases suggest that dietary patterns can potentially significantly lower the risk of certain diseases such as cancer, cardiovascular disease, and Alzheimer's disease [44,45,46,47,48,49]. Currently, diets enriched in natural antioxidants, vitamins, and phytochemicals are perceived as healthful by Western countries, which use such nutritional supplements regularly [18]. Yet, a limited understanding persists about how dietary compounds actually can affect human metabolism. Even essential nutrients, while necessary at certain levels, can be harmful at high doses, leading to hepatotoxicity and DNA damage [50].
Similarly, while moderate consumption of some food items could have protective effects, higher intake could lead to health problems [51,52,53,54]. For example, moderate coffee consumption has been linked to a lower risk of coronary heart disease, which is probably due to antioxidants found in coffee. High coffee consumption, however, has been reported to increase cholesterol levels, which is probably due to the high levels of diterpenes present in unfiltered coffee [51,52,53,54]. Moreover, nutrigenomics and nutrigenetics research shows that genetic variations could alter the absorption, metabolism, excretion, and biological response to food-derived components of individuals [53,54]. For example, specific genetic polymorphisms have been associated with a slow rate of caffeine metabolism, which may also affect health [51,52]. Such evidence suggests that a combination of genomics and metabolomics might result in optimal nutritional recommendations for individuals.
The Cellular Functions of Food Components
Food-derived metabolites interact with genes, proteins, enzymes, and microenvironment, affecting cell metabolism through three major mechanisms.
First, metabolites constitute the building blocks of large macromolecules (e.g., DNA, proteins, and oligosaccharides) and cellular membranes (e.g., lipid bilayers). For that reason, the bioavailability and relative composition of the various metabolites directly determine the chemical-physical properties of such large macromolecules. For example, it has been shown that animals fed a diet enriched with particular fatty acids could alter the composition of their neural membranes [55,56,57]. The altered composition affects the membranes' shape and flexibility, the function of intramembrane proteins such as ion channels, and, ultimately, neurotransmission and brain development [55,56,57].
Second, dietary metabolites can provide a source of energy or regulate pathways for energy metabolism, or both. For example, when energy is needed, dietary sugars can readily enter glycolysis as substrates, leading through the citric acid cycle and oxidative phosphorylation pathways to the generation of energy-rich metabolites such as nucleotide adenosine triphosphate. When there is an excess of adenosine triphosphate in the system, the metabolism of dietary sugars is promptly directed toward the pathway for fatty acid synthesis. Recent research, indeed, provides compelling evidence that foods and beverages rich in sugar, such as high-fructose corn syrup, lead to fatty liver and diabetes and eventually chronic inflammatory diseases [58]. Alternatively, other dietary metabolites, such as B-complex vitamins, might bind to enzymes as cofactors modifying their catalytic activities or assisting in their functions. For example, the cofactor thiamine pyrophosphate is a thiamine (vitamin B1) derivative that catalyzes several biochemical reactions including the activity of the multi-enzyme, complex pyruvate dehydrogenase at the junction of glycolysis and the citric acid cycle. Likewise, biotin (vitamin B7) is required for acetyl-coenzyme A (CoA) carboxylase activity for the synthesis of fatty acids.
Third, dietary metabolites can act as signaling messengers. For example, fatty acids metabolites can bind and activate cytosolic/nuclear receptors or membrane-bound G protein-coupled receptors, transducing biochemical signals. Alternatively, metabolites can modify nucleic acids (e.g., DNA), proteins (e.g., histones), and enzymes (histone deacetylases) [59,60]. When they do so, they modulate gene expression and even reprogram our genetic code. The most common of such so-called ‘epigenetic modifications' include enzymatic addition of acetyl or methyl groups. These modifications are regulated by the availability, subcellular compartmentalization, and concentration of metabolites such as acetyl-CoA and S-adenosylmethionine. Recently, it has been shown that fluctuation in the levels of energy metabolites such as nicotinamide adenine dinucleotide (NAD+) and the ketone body beta-hydroxybutyrate associated with circadian rhythms or particular diet patterns (e.g., fasting, calorie restriction, and low carbohydrate diets) can be responsible for the modulation of enzyme activities and gene expression, ultimately leading to an increase in life span in animal models [59].
In addition to the three general mechanisms mentioned previously, some dietary metabolites modulate cell metabolism by acting as antioxidants, scavenging the cell from the damage caused by oxidation. Dietary antioxidants can interact with free radicals, peroxides, metals, and oxygen. In doing so, they inhibit the formation of reactive oxygen species or the propagation of oxidative stress signaling that could damage all components of the cell, including proteins, lipids, and DNA. Naturally occurring antioxidants are retinoids (vitamin A), tocopherols (vitamin E), and ascorbic acid (vitamin C). These metabolites, together with some man-made antioxidants such as butylated hydroxytoluene, butylated hydroxyanisole (BHA), and propyl gallate are also artificially added to food products to stop them from going rancid and preserve their taste.
In summary, a better understanding of the effects of dietary metabolites at the cellular level on genes, proteins, enzymes, and microenvironments might allow a rational design of diets for the purpose of manipulating cell functions and enhancing overall health.
Dietary Lipids and Lipidomics
Together with proteins and carbohydrates, fats constitute one of the largest classes of nutrients. Lipids, a subgroup of metabolites mostly insoluble in water, provide the bulk of the fats. Classical examples of dietary lipids are cholesterol, triglycerides, saturated fatty acids, and trans fat. Their intake has been linked to various pathological conditions including obesity, metabolic syndrome, and cardiovascular diseases [61]. Not all lipids, however, have been associated with an increased risk of diseases. Indeed, some other well-known dietary lipids, such as omega-3 fatty acids, have been linked to human health, which might explain their exponential growth in nutraceuticals as dietary supplement or functional food (Boxes 1-3) [62,63,64]. Because lipids are involved in crucial biological mechanisms, there is a growing public and scientific interest in understanding their role in nutrition.
Box 3. The complex metabolism of omega-3 fatty acids

The metabolomic analysis of lipids is called lipidomics [65]. Although lipidomics falls under the umbrella of metabolomics, the distinct solubility properties of lipids often dictate their separate analysis in metabolomic experiments. Indeed, contrary to polar metabolites such as amino acids and nucleotides, lipids are mostly insoluble in water and must be extracted from biological samples using organic solvents or distinct, solid-phase extraction procedures. Lipidomic approaches can be applied for the development of appropriate diagnostic tests to monitor nutritional unbalances or deficiencies in dietary lipids such as the ratio of omega-6 versus omega-3 fatty acids (Box 2).
Microflora and Regional Metabolic Phenotypes
Diets modulate the community of microorganisms in the human digestive system [66]. Food may indeed contain microorganisms or provide nourishment for their proliferation and activity in the gut. Each human carries a complement of at least 160 bacterial species, with more than 536,000 bacterial genes between them - more than 20 times the human genome. Such microorganisms are often essential for optimal nutrient absorption, bioavailability, and metabolism [67]. Products of microbial metabolism enter the host metabolome, causing substantial variability in the metabolomic profiles [19]. Some of the microbial-derived metabolites have been shown to be beneficial for health and the immune system; others have been linked to chronic inflammatory diseases [67,68]. The use of nutraceuticals such as probiotics (live microorganisms) and prebiotics (non-digestible food ingredients such as polysaccharides) to modulate the growth or activity of gut bacteria has been proposed as medical strategy to prevent or treat diseases (Box 1) [69,70].
Along with common diets, medicinal practices, genetics, and other lifestyle and environmental factors, the gut microbes strongly contribute to regional metabolomic phenotypes [1,67,71]. A study of individuals living in Japan, Northern China, Southern China, and the West (US and UK) reported significant differences between the urinary metabolic profiles based on geography [1,67]. The characterization of regional or individual microbiomes and their role in the absorption and metabolism of food-related compounds will provide valuable insight for more tailored nutrition [72].
Multi-Organ Systems Metabolomics
The metabolism of dietary components in the human body often occurs in multiple organs. Following their ingestion, many of the components of food are absorbed in the gastrointestinal tract by the microbiome, enterocytes, or both. Most of these components or their metabolites are then transported into the liver, where they can undergo further modification before appearing in the plasma for delivery to the various tissues of the body. Different cell types can metabolize food components in various ways. Later, these compounds may appear, with or without chemical modification, in the urine.
Thus, a combination of dietary intake, gut absorption, liver metabolism, transport, and further chemical modifications often contributes to the regulation of dietary components in humans. A holistic, multi-organ system approach aiming to screen metabolites in various tissues of the body is often required to understand the metabolism of a particular metabolite in humans. An example of such an approach is described in Box 3 [73].
Notably, it has been reported that genetic polymorphisms could contribute to the absorption, transport, metabolism, and biological effects of dietary omega-3, explaining some of the inconsistencies in previous studies relating polyunsaturated fatty acid (PUFA) intake and health benefits [74,75,76,77,78]. In particular, some of the effects of omega-3 supplementation seem to be related to the ApoE polymorphism [75,77,78]. Such evidences highlight the importance to integrate metabolomics and lipidomics studies into those from other -omics techniques including genomics, transcriptomics, and proteomics in order to obtain a more accurate interpretation and classification of the data sets [71].
Metabolomics Tools and Strategies
Innovative technologies are rapidly facilitating our ability to measure the numerous and diverse metabolites in biological samples. One of the main challenges for metabolomics is the generation of comprehensive profiles of metabolites in biological samples [79]. Metabolites vary in concentrations (from attomolar to millimolar), chemical complexity (thousands of components), and spatial localization. Complex analytical strategies have been designed to study metabolic phenotypes as well as to perform comparative analyses of metabolomes. Currently, three main strategies are used for metabolomic investigations: untargeted metabolomics, targeted metabolomics, and in situ metabolomics.
Untargeted Metabolomics
Untargeted metabolomics consists of the unbiased screening of metabolites in a biological sample (fig. 4). The intention of this approach is to compare patterns or ‘profiles' of metabolites among different sample groups. The untargeted metabolomics approach is used in nutrition research to screen for molecular composition of food, characterize the metabolic phenotypes of individuals, and determine the outcome of dietary interventions on human metabolism.
Untargeted metabolomics. An untargeted approach allows the identification of alterations in metabolic profiles induced by a disease state or nutritional intervention. Usually, liquid chromatography tools are used to separate and screen complex mixtures of metabolites extracted from biological samples. In this example, metabolites were separated using UPLC coupled with a hybrid QToF system mass spectrometer [80,81]. The analysis provided a metabolite profile, which is a biochemical snapshot of the metabolite inventory of the tissue under investigation. Metabolite differences between groups can be analyzed using informatics solutions, which provide multivariate statistical analyses tools and database searches functionalities (e.g., METLIN, Human Metabolome Database, and LipidMaps) for the identification of the metabolites.
Untargeted metabolomics. An untargeted approach allows the identification of alterations in metabolic profiles induced by a disease state or nutritional intervention. Usually, liquid chromatography tools are used to separate and screen complex mixtures of metabolites extracted from biological samples. In this example, metabolites were separated using UPLC coupled with a hybrid QToF system mass spectrometer [80,81]. The analysis provided a metabolite profile, which is a biochemical snapshot of the metabolite inventory of the tissue under investigation. Metabolite differences between groups can be analyzed using informatics solutions, which provide multivariate statistical analyses tools and database searches functionalities (e.g., METLIN, Human Metabolome Database, and LipidMaps) for the identification of the metabolites.
A typical untargeted experiment starts with the extraction of the metabolites from the biological samples. Next, the components are separated using chromatographic techniques such as liquid chromatography and gas chromatography or directly infused in a mass spectrometry (MS) detector [80,81]. Mass spectrometers that offer high resolution and high accuracy are generally used, including the time of flight, Fourier transform ion cyclotron resonance, and orbitrap.
Dealing with variations in thousands of molecular species, the untargeted metabolomics strategy relies on statistical tools such as computational sorting as well as multivariate and pattern-recognition analyses to group the observed changes in metabolites. Although statistical approaches could provide important information on the actual metabolite species (e.g., mass and retention time in LC-MS experiments), additional investigative work using MS/MS information [81,82,83], metabolite databases, and confirmatory analysis using standards is required to be fully confident of the assignment (fig. 5).
Structural elucidation. The fine chemical structure of complex metabolites can be characterized using modern tools based on MS [81,82,83]. a Applying high collision energy (MS/MS), complex structures of precursor molecules (MS) can be broken down into constituent parts, from which one can deduce the original structure. In this example, the identification of a phosphatidylcholine molecule is based on the observation of a characteristic fragment (phosphocholine) produced upon collision induced dissociation (b) using a hybrid QToF system [81,82].
Structural elucidation. The fine chemical structure of complex metabolites can be characterized using modern tools based on MS [81,82,83]. a Applying high collision energy (MS/MS), complex structures of precursor molecules (MS) can be broken down into constituent parts, from which one can deduce the original structure. In this example, the identification of a phosphatidylcholine molecule is based on the observation of a characteristic fragment (phosphocholine) produced upon collision induced dissociation (b) using a hybrid QToF system [81,82].
Targeted Metabolomics
Targeted metabolomics focuses on analyzing selected metabolites, often related to a specific metabolic pathway (e.g., fatty acids, oxylipins, amino acids, acylcarnitines, or particular classes of phytochemicals) (fig. 6) [84,85,86,87,88,89,90]. In most cases, targeted metabolomics is a hypothesis-driven approach. The metabolites for analysis are selected according to the questions asked, and specific analytical methods are developed for their absolute quantification. This approach can be used in nutrition science to determine information on concentration, bioavailability, turnover, or metabolism of dietary compounds.
Targeted metabolomics. Targeted approaches can be used to validate the observed alterations in metabolic profiles induced by disease status or nutritional intervention. Furthermore, they can be used to quantify low-abundance bioactive metabolites such as prostaglandins and other oxygenated PUFA derivatives. In this example, omega-3 metabolites were detected using UPLC in combination with a tandem quadrupole [84,85].
Targeted metabolomics. Targeted approaches can be used to validate the observed alterations in metabolic profiles induced by disease status or nutritional intervention. Furthermore, they can be used to quantify low-abundance bioactive metabolites such as prostaglandins and other oxygenated PUFA derivatives. In this example, omega-3 metabolites were detected using UPLC in combination with a tandem quadrupole [84,85].
A typical targeted experiment starts with the extraction and purification of the selected metabolites from the biological samples. Next, the components are separated using liquid chromatography or gas chromatography before MS detection. Technological advancements contribute to increasing the number of metabolites that can be quantified simultaneously in a single analysis, permitting monitoring of several hundred metabolites in a single analysis. Today, using selected retention times with ultra-performance liquid chromatography (UPLC) and multiple-reaction monitoring transitions with triple-quadrupole instruments, hundreds of metabolites can be monitored simultaneously [90].
An example of a targeted metabolomics approach is illustrated in figure 6 for the study of the metabolism of omega-3 fatty acids. Omega-3 fatty acids are produced through a sequential biochemical pathway that yields a series of functionally distinct signaling lipids, each of which could further generate bioactive metabolites [91] through different biochemical pathways. The simultaneous quantification of each of the low-abundance molecular species can be used to shed light on the regulation of omega-3 biochemical pathways (fig. 6).
In situ Metabolomics
In situ metabolomics approaches provide the detailed spatial distribution of metabolite species on a tissue, a new level of description beyond the pure measure of metabolite concentration. Metabolites, indeed, are localized in different compositions and concentrations within tissues and even cell compartments. Such a level of information is often missed using traditional sample preparation and metabolite extraction protocols for metabolomic analysis.
There are two main biological applications for in situ metabolomics: MS imaging and real-time MS. MS imaging involves the irradiation of a frozen tissue section with a focused excitatory beam - a laser, ions, charged droplets of solvent, for example. With a spatial resolution of 1-200 μm, the beam scans through tissues along all three axes [92,93,94]. Upon impact, the sample surface releases a vapor of ionized molecules that can be directed into the detector for the purpose of measuring accurate masses and thus generating topographic maps of metabolite composition. MS imaging generates spectacular images of the molecular composition of tissues, providing a spatial distribution of metabolites in their natural environment. MS imaging can be used in nutritional research to investigate the absorption, distribution, and metabolism of food-derived components and their effects on physiology (fig. 7) [92,93,94,95,96,97].
MS imaging. Scanning sections of biological tissues along the three axes with a laser allows the ionization of the constituents and their detection by MS. Such information can be represented as topographical maps of molecular composition. In this example, a functional MS imaging using MALDI-Synapt allows to determine the exact localization in the rat brain of the changes in molecular composition induced by a particular diet [92,93,94,102].
MS imaging. Scanning sections of biological tissues along the three axes with a laser allows the ionization of the constituents and their detection by MS. Such information can be represented as topographical maps of molecular composition. In this example, a functional MS imaging using MALDI-Synapt allows to determine the exact localization in the rat brain of the changes in molecular composition induced by a particular diet [92,93,94,102].
Real-time MS allows for real-time, rapid in situ screening of food and biological tissues (fig. 8). Many new desorption ionization technologies have been applied to this kind of metabolomics analysis [96,98], including direct analysis in real time (fig. 8) [35], desorption electrospray ionization, and atmospheric solids analysis probe [99].
Real-time MS. Noveldesorption ionization tools allow the real-time analysis of food and biological samples, which could be used for quality assessment, traceability, and diagnosis. In this example, human sebum and fish oil are analyzed for their molecular content using a novel technological solution, direct analysis in real time (IonSense, Saugus, Mass., USA), in combination with ion-mobility separation of a Synapt G2-S HDMS system (Waters Corp, Milford, Mass., USA). Samples were swiped on a capillary and placed near the ion source of the mass spectrometer and then separated by ion-mobility MS. Software solutions allow the automatic detection of differences in PUFA composition.
Real-time MS. Noveldesorption ionization tools allow the real-time analysis of food and biological samples, which could be used for quality assessment, traceability, and diagnosis. In this example, human sebum and fish oil are analyzed for their molecular content using a novel technological solution, direct analysis in real time (IonSense, Saugus, Mass., USA), in combination with ion-mobility separation of a Synapt G2-S HDMS system (Waters Corp, Milford, Mass., USA). Samples were swiped on a capillary and placed near the ion source of the mass spectrometer and then separated by ion-mobility MS. Software solutions allow the automatic detection of differences in PUFA composition.
Conclusions and Future Directions
The recent interest in applying metabolomics for nutrition science coincides with a shift in the medical community and general population toward disease prevention and treatment through adequate food intakes and diets. By offering a snapshot of the molecular composition of food as well as the individual's nutrition and health status, metabolomics is set to provide valuable information to health-care professionals in terms of diagnosis and diet counseling. Metabolomics promises to identify individual variations in dietary requirements classifying individuals into specific groups based on their ‘metabotype'. Eventually, such a strategy could lead to the development of ‘personalized nutrition', in which diet is attuned to the nutritional needs of individual patients [72,100]. Specific blood-metabolite profile tests might one day identify persons with specific dietary deficiency or who are at risk for disease. Based on individual genetic variations, personalized dietary recommendations and supplements may be advised for such individuals, the aim being not merely to decrease the risk of disease but to achieve optimal health and wellness [6,101].
Acknowledgements
G.A. is grateful to the Alzheimer's Association grant (NIRG-11-203674) for the financial support.





![Fig. 4. Untargeted metabolomics. An untargeted approach allows the identification of alterations in metabolic profiles induced by a disease state or nutritional intervention. Usually, liquid chromatography tools are used to separate and screen complex mixtures of metabolites extracted from biological samples. In this example, metabolites were separated using UPLC coupled with a hybrid QToF system mass spectrometer [80,81]. The analysis provided a metabolite profile, which is a biochemical snapshot of the metabolite inventory of the tissue under investigation. Metabolite differences between groups can be analyzed using informatics solutions, which provide multivariate statistical analyses tools and database searches functionalities (e.g., METLIN, Human Metabolome Database, and LipidMaps) for the identification of the metabolites.](https://karger.silverchair-cdn.com/karger/content_public/journal/jnn/6/4-5/10.1159_000354403/2/m_000354403_f04.gif?Expires=1716301488&Signature=G4tY8KmWThVLUhOVxxs3y4lcuPZIMGN8437Obv~haS4QabENcBCqdx3fy1iECO~Oa4g~vZn0y3ptDPso01tM0GcM3Zq~bR8jjEYSaxgNxj0~6LejFah-ZmnmLTcVPMSn2Iv4fTvMda2Gp98U-gLnK402lxTrfTTOseImS0XTxQlaL5uPtWPT3Yi~daUhxRtZtsQgNi80d2y8PdSJaxwXJ8FhKjos8TWQbKMJhpLlAUY3pLwEv9Oc4LcSF1riyvHfRzkAdSYgZsmG~7VLhcBNGdaT2MZE9baexJQfmSjYGUkp0n5dPPqhOQIcbl6NsBh-NKoFOBTmi6uqN5vMZDz57g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. Structural elucidation. The fine chemical structure of complex metabolites can be characterized using modern tools based on MS [81,82,83]. a Applying high collision energy (MS/MS), complex structures of precursor molecules (MS) can be broken down into constituent parts, from which one can deduce the original structure. In this example, the identification of a phosphatidylcholine molecule is based on the observation of a characteristic fragment (phosphocholine) produced upon collision induced dissociation (b) using a hybrid QToF system [81,82].](https://karger.silverchair-cdn.com/karger/content_public/journal/jnn/6/4-5/10.1159_000354403/2/m_000354403_f05.gif?Expires=1716301488&Signature=afG-x63YzBFI5zbOjxbG6T899ZtlTR2JjxCO3UMaKGF3pNvb7r01IGOOOfOJinfj6O0b524uN6EWc87V-h-lQWkIo4~rtNErz3xjqUX9-cpWbJuUXblTrLH5KGwxEsldw-QFPA~J~eNmOXNu7whbwl0uNcsapJPYKr7w03mu5XmZOcAtGrHR0u9N8BGKHvgKhbKc0urfiAEjpg608c-xbc~W852VT6xmxWwyLFfZRiw0wz7QVvy8JAoFrZteZCfxvwP3T6QL4fzbF5C9nc3bzoZ0ZYoJzNRKji7SIp2RQPeIBSfz~CVYNzREPZ0~b4ALZxZEQj~bvt6NaEKQnwc4Bw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 6. Targeted metabolomics. Targeted approaches can be used to validate the observed alterations in metabolic profiles induced by disease status or nutritional intervention. Furthermore, they can be used to quantify low-abundance bioactive metabolites such as prostaglandins and other oxygenated PUFA derivatives. In this example, omega-3 metabolites were detected using UPLC in combination with a tandem quadrupole [84,85].](https://karger.silverchair-cdn.com/karger/content_public/journal/jnn/6/4-5/10.1159_000354403/2/m_000354403_f06.gif?Expires=1716301488&Signature=ok3XHtFBKQMGNLkfxUyRYThUIlgO9eFVVzSAw8BS8EIPYokLiJlyo4NaVxEkjmF1YGOTecEHf0m2WRYY7wonskrGYp4ME0wYfp8VH~LLVhXP-CipBZ0FxOHEgf575rx7i58n8hZSuRsw32I46DWNaJQ~u99Ztvi2pAwBeSqVgE1H9n4Ihpxkp8j2U84nP2RLQwv8R5FpyqFeEKIkb08emQr5mY7f4efDDHiZn~pukjyPwItxtoJo71h~Tol02z3uNLis03QSUuVH40kw1OkpmEyW-qMz~qGDXFDRcKc-ULnlwoM2HC2YqDToqfiD2nzF5FtcyQDiNLJ3iw2YIqGEvg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 7. MS imaging. Scanning sections of biological tissues along the three axes with a laser allows the ionization of the constituents and their detection by MS. Such information can be represented as topographical maps of molecular composition. In this example, a functional MS imaging using MALDI-Synapt allows to determine the exact localization in the rat brain of the changes in molecular composition induced by a particular diet [92,93,94,102].](https://karger.silverchair-cdn.com/karger/content_public/journal/jnn/6/4-5/10.1159_000354403/2/m_000354403_f07.jpeg?Expires=1716301488&Signature=fGisFBJndwYmAylmqk3zyJJ848gdFRhpDJzcNrzunZW~yPWUM0teNa4NUabAjApbOgYL57yXfFFkkJxGmdXy4laUeDoF6XZ-OURr0cmkB7CLRpXpXc1U1YCkZCqy5uHcPapMejbo9odH0gp8pmOnf7kzp6xLSde5zImihEDZOVqDK0lyJefzNtWq7b0PTlnkIIQXYds-dZKbVLwTjz~xUcl3ZYWe0eZylRF~PXhQeQViwcB179DftlVa553W4M7dfied4rqDymQafqo--miujS~ujjnYtobkVi2UWvOZk0MIK4xgQRFOZE9y2eKJQrNBFh-Qt2GIRjWUEUksTH4mdw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
