Abstract
MicroRNAs (miRNAs) are a family of highly conserved noncoding single˗stranded RNA molecules of 21 to 25 nucleotides. miRNAs silence their cognate target genes at the post-transcriptional level and have been shown to have important roles in oncogenesis, invasion, and metastasis via epigenetic post-transcriptional gene regulation. Recent evidence indicates that the expression of miR-181a is altered in breast tumor tissue and in the serum of patients with breast cancer. However, there are several contradicting findings that challenge the biological significance of miR-181a in tumor development and metastasis. In fact, some studies have implicated miR-181a in regulating breast cancer gene expression. Here we summarize the current literature demonstrating established links between miR-181a and human breast cancer with a focus on recently identified mechanisms of action. This review also aims to explore the potential of miR-181a as a diagnostic and/or prognostic biomarker for breast cancer and to discuss the contradicting data regarding its targeting therapeutics and the associated challenges.
Introduction
MicroRNAs (miRNAs) are a group of endogenous small non-coding regulatory RNAs of 21 to 25 nucleotides. miRNAs silence their target genes at the post-transcriptional level by inhibiting mRNA translation or by degrading mRNA molecules through binding to their 3’-untranslated regions (UTR) [1]. Many of these target genes are essential for regulating fundamental biological processes such as cell proliferation, differentiation, apoptosis, and migration [1, 2]. miRNAs have been shown to play an important role in oncogenesis, invasion, and metastasis. In addition, altered expression of miRNAs has been demonstrated in a variety of cancer states [3-5].
Members of the miRNA-181 (or miR-181) family are evolutionarily conserved across almost all vertebrates, suggesting their functional importance. The miR-181 family contains four immensely conserved mature members (miR-181a, miR-181b, miR-181c, and miR-181d) [6, 7]. Human miR-181a and miR-181b genes are clustered together and located on two chromosomes, 1 and 9, whereas miR-181c and miR-181d genes are clustered together on chromosome 19 [8, 9]. Recent studies have reported the involvement of miR-181a in diverse cellular functions such as growth, proliferation, death, survival, and maintenance, as well as tumor suppression and carcinogenesis [10, 11]. Though some studies have shown that miR-181a expression is downregulated in several human solid tumors [12-17], others have demonstrated that upregulation of miR-181a may promote metastasis and invasion of human cancers [18-20].
Breast cancer is the most common malignant tumor in females. It represents a complex heterogeneous group of tumors that display significant diversity with respect to histopathological features and therapeutic responses [21]. Tumor grade and hormone receptor status, including that for estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor (EGF)-like receptor 2 (HER2) are often used clinically for breast cancer classification and treatment selection [22]. Survival from this disease depends on the stage at diagnosis, with patients who are detected and diagnosed early having more favourable outcomes. Thus, many researchers have focused on biomarkers to facilitate detection of breast cancer in its early stages; miRNAs have shown immense potential for this purpose [23]. In fact, a growing body of evidence has linked the pathological role of miR-181a to breast cancer, and the expression of miR-181a has been found to correlate with clinicopathological features of breast tumors [24-28]. In this review, we will discuss the differing oncogenic roles of miR-181a and its potential as a biomarker and pharmacologic target for the treatment of metastatic breast cancer.
Anti-oncogenic Roles of miR-181a
Several recent studies have demonstrated that miR-181a exerts anti-breast cancer effects by preventing tumor invasion and metastasis, reducing mammosphere formation, inducing cancer cell death, and enhancing drug sensitivity. In other studies, miR-181a was downregulated in more aggressive or late stage breast cancer and therefore appeared to function as a tumor suppressor gene (Table 1).
Summary of the relative expression of miR-181a in breast cancer tumor tissue and cell lines. The relative expression of miR-181a in breast tumor tissue or cell line in the literature is grouped with the relevant references (in parentheses). Abbreviations: ER, estrogen receptor; TNBC, triple negative breast cancer; TNFα, tumor necrosis factor alpha; TGF-β, transforming growth factor beta

miR-181a expression is downregulated in aggressive or late stage breast cancer
miR-181a expression in breast tumor tissue. Triple negative breast cancer (TNBC) is defined as ER-negative (ER-), PR-negative (PR-), and HER2-negative (HER2-) [29]. Patients with TNBC have low survival rates. TNBC is classified into six molecular subtypes: basal-like 1 and 2, immunomodulatory, mesenchymal, mesenchymal stem-like, and luminal androgen receptor subtype [30]. In TNBC tumor samples (n=32), miR-181a was downregulated more than 1.5-fold compared to its expression in corresponding normal breast tissue [31]. Moreover, lymph node involvement in breast cancer is indicative of high-grade, late-stage advanced disease. In breast cancers where a lymph node could be identified using realtime RT-PCR (n=31), miR-181a was one of the eight miRNAs that was downregulated when compared to the non-lymph node (n = 42) group [32].
miR-181a expression in breast cancer cell lines. In human breast cancer cell lines, miR-181a expression is significantly lower in aggressive cancer cells such as TNBC adenocarcinoma MDA-MB-231 and breast cancer stem SK-3rd cells as compared to less aggressive breast cancer cell lines such as MCF-7 (ER+/PR+/HER2-; one of the most low-invasive breast cancer cells) and SK-BR3 (ER-/HER2+) [33]. Downregulation of miR-181a is also observed in multidrug-resistant MCF-7 cells compared with its parental cell line [19], and in mitoxantrone-resistant MCF-7 cells [34]. The MCF-7 cell line retains several characteristics of differentiated mammary epithelium, including the ability to process estradiol (E2) via cytoplasmic ER and the ability to form domes. Estrogen functions as a mitogen in ER-positive (ER+) MCF-7 cells. The expression of miR-181a is suppressed by E2 or tumor necrosis factor-alpha (TNFα), and the combination of E2 and TNFα results in further suppression [25, 35]. Taken together, the downregulation of miR-181a in these breast cancer cells suggests that it may function as an anti-oncomir.
miR-181a targets specific genes to inhibit breast cancer
miR-181a prevents breast cancer metastasis. Matrix metalloproteinase (MMP)-14 is a key proteolytic enzyme involved in the degradation of the extracellular matrix. It is highly expressed in aggressive breast tumors and is linked to enhanced cancer invasion [36]. Li et al. established that in breast tumors, MMP-14 expression is regulated by miR-181a through a posttranscriptional mechanism. They first validated a miR-181a response element within the 3’-untranslated region of MMP-14, and then showed the reduction of MMP-14 expression by ectopic miR-181a. Moreover, in support of a critical relationship between these two genes, miR-181a–mediated reduction of MMP-14 levels was sufficient to decrease cancer cell migration and invasion in in vitro and in vivo assays [33].
In another study, Noh et al. suggested that elevation of miR-181a levels may prevent breast cancer metastasis [37]. They investigated the effect of impaired miRNA processing on invasion-essential urokinase-type plasminogen activator (uPA) expression in breast cancer cells. Drosha, DGCR8, and Dicer, are important components of the miRNA processing machinery. uPA is a serine protease recognized as a key molecule in cancer invasion and metastasis [38-40]. Noh et al. found that the level of mature miR-181a was lower in high uPA-expressing cancer cells compared to cells with low uPA expression [37]. Knockdown of Drosha, DGCR8, or Dicer substantially increased in vitro invasion of cancer cells and elevated uPA levels resulting from less mature miR-181a. Importantly, uPA mRNA was identified as a direct target of miR-181a, and the reduced expression of uPA was observed in TNBC cell lines (MDA-MB-231 and MDA-MB-436) [37].
Nonetheless, Kastrati et al. demonstrated that miR-181a is capable of impairing ER+ mammosphere formation by directly targeting Pleckstrin Homology-Like Domain, Family A, member 1 (PHLDA1) [35]. PHLDA1 is essential for the formation of ER+ mammospheres, and its elevated expression is associated with a higher risk of distant metastasis in patients with ER+ breast cancer [35]. In ER+ breast cancers, upregulation of PHLDA1 in mammospheres is largely dependent on crosstalk between the ER and inflammatory nuclear factor kappa B (NFκB) pathway.
miR-181a promotes breast cancer cell apoptosis. miR-181a can promote the efficacy of adriamycin-based neoadjuvant chemotherapy by enhancing adriamycin-induced apoptosis of MCF-7 cells [19]. B-cell lymphoma 2 (Bcl-2) has an inhibitory effect on mitochondrial metabolism and adriamycin-induced apoptosis in MCF-7 cells. Zhu et al. demonstrated that miR-181a decreased the expression of Bcl-2 by forming imperfect base pairing with the 3’-UTR of the Bcl-2 gene. As a result, a negative relationship between miR-181a and Bcl-2 in MCF-7 and multidrug-resistant MCF-7 cells was observed [19]. Moreover, miR-181a has been shown to promote anoikis in a human mammary epithelial cell line (MCF10A) [41]. Anoikis is a form of apoptosis induced by insufficient cell-matrix interactions and a critical player in tumor angiogenesis and metastasis [42]. Wei et al. demonstrated that miR-181a promotes anoikis by inhibiting the expression of autophagy-related protein ATG5 in MCF10A cells [41].
miR-181a enhances drug sensitivity. The human breast cancer resistance protein (BCRP/ ABCG2) is the second member of the G subfamily of the large ATP-binding cassette (ABC) transporter superfamily. BCRP was initially discovered in multidrug resistant breast cancer cell lines where it confers resistance to chemotherapeutic agents such as mitoxantrone. miR-181a enhances drug sensitivity in mitoxantrone-resistant breast cancer cells through targeting BCRP [34]. Jiao et al. showed that overexpression of miR-181a downregulates BCRP expression via binding to the 3’-UTR of BCRP mRNA, and sensitizes mitoxantrone-resistant MCF-7 cells to mitoxantrone. Thus, they proposed that miR-181a may be a potential target for preventing and reversing drug resistance in breast cancer [34].
Oncomir Activities of miR-181a
Despite available evidence in favor of an anti-oncomir role for miR-181a, some emerging studies suggest an oncogenic role in breast cancer. High miR-181a levels were associated with poor survival rates after treatment in patients with breast cancer [28, 43]. miR-181a overexpression has been linked to aberrant activation of major pathways involved in breast tumorigenesis, including transforming growth factor-β (TGF-β) [18, 24]. Two independent research groups have reported that TGF-β upregulates the miR-181a levels in breast cancer cells [18, 44]. They demonstrated that TGF-β upregulates members of the miR-181 family (including miR-181a) at the post-transcriptional level through Smad4-independent functions of Smad2/3. TGF-β induces binding of Smad2/3 to the primary transcripts of miR-181a and regulates their maturation. Furthermore, their data suggest that miR-181a is required for TGF-β-mediated breast cancer cell migration and invasion.
miR-181a expression is upregulated in TNBC and aggressive breast cancer
Upregulation of miR-181a expression was found in TNBC by three independent research groups where the increased expression of miR-181a correlated with breast cancer aggressiveness [24, 26, 45] (Table 1). Bisso et al. investigated the expression of miR-181a in a panel of 104 primary breast cancers and eight normal tissue counterparts. The expression of miR-181a was upregulated in tumor specimens when compared with normal tissue, and, importantly, the expression levels correlated positively with tumor aggressiveness, with grade 3 tumors showing the highest expression of miR-181a compared to grade 1 and 2 tumors [27]. The clinicopathologic significance of miR-181a accumulation in bone marrow was investigated by Ota et al. in 291 patients with breast cancer. In this study, significantly more cases with high levels of miR-181a were grade 3, hormone receptor–negative, HER2-positive, and experienced cancer recurrence compared to cases with low miR-181a levels [28]. Moreover, high levels of miR-181a were detected in bone marrow obtained from breast cancer patients with recurrent disease but not in patients without recurrence [28]. Invasive micropapillary carcinoma is an uncommon histological type of breast cancer with an aggressive behavior relative to invasive ductal carcinomas of no special types [46]. Upon investigation of miRNA expression in invasive micropapillary carcinoma samples (n = 22) and invasive ductal carcinomas of no special types (n = 24) via RT-qPCR, Li et al. found higher levels of miR-181a in invasive micropapillary carcinoma [46]. These findings suggest that miR-181a is a predictive biomarker for breast cancer metastasis and patient survival.
Targeting specific genes in breast cancer
miR-181a promotes breast cancer cell survival and metastasis. There are several independent studies that support the contribution of miR-181a to breast cancer survival and metastasis. Firstly, miR-181a was found to directly target the tumor suppressor gene, programmed cell death protein 4 (PDCD4) [28]. PDCD4 is widely recognized as an important tumor suppressor, which is an inhibitor of tumor neoplastic transformation. Loss of PDCD4 expression has been observed in advanced primary breast cancer cases [47]. Secondly, miR-181a was reported to induce sphere formation and reduce DNA damage response (DDR) by downregulating the ataxia telangiectasia mutated (ATM) gene [18, 27]. The ATM gene encodes a large protein that belongs to a kinase family. Members of this family have been shown to repair DNA at cell cycle checkpoints following DNA damage [48]. DDR activation occurs at early stages of transformation and results from oncogene deregulation. The activation of ATM-regulated DDR signaling may delay or prevent cancer [49, 50]. miR-181a functions as a negative regulator of DDR in breast cancer through impacting the expression and activity of the ATM or its substrate checkpoint kinase 2 [27, 51]. Thirdly, dysregulated TGF-β signaling is involved in late-stage breast cancer metastasis. Taylor et al. established that miR-181a is a TGF-β-regulated “metastamir” that enhanced the metastatic potential of breast cancers by promoting epithelial-mesenchymal transition, migration, and invasive phenotypes. Mechanistically, miR-181a could promote anoikis resistance through directly targeting pro-apoptotic factor Bim [24].
miR-181a elevates chemotherapeutic resistance. Acquired therapeutic resistance is a major drawback to effective systemic therapies for cancer. TNBCs are the most aggressive breast cancer subtype characterized by high rates of relapse, visceral metastasis, and early death [52]. Aggressive TNBCs quickly develop resistance to chemotherapy. Niu et al. reported that induction of miR-181a by genotoxic treatments promotes chemotherapeutic resistance and metastasis in breast cancer [45]. After doxorubicin treatments, the increased expression of miR-181a was detected in TNBC cells including MDA-MB-231, HCC1937, and HER2+ BT474. In this study, the authors identified BAX as a direct functional target of miR-181a whose suppression decreased apoptosis and increased invasion of TNBC cells upon doxorubicin treatment. Suppression of pro-apoptotic factor BAX by miR-181a resulted in decreased apoptosis and increased invasion of TNBC cells upon doxorubicin treatment.
Summary of the Diverse Roles of miR-181a in Breast Cancer Progression
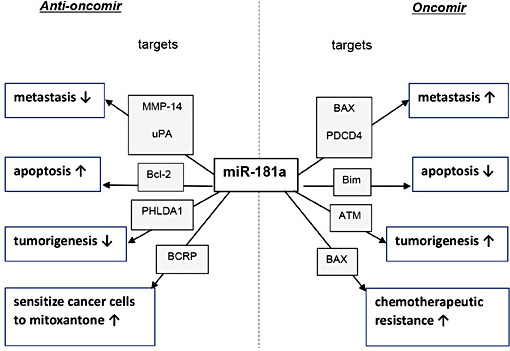
So far, we have reviewed contradictory reports regarding the expression and role of miR-181a in breast cancer (Table 1 and Table 2). Some data support miR-181a as an oncogene [18, 24, 27, 28, 45], whereas other data suggest a tumor suppressor function [19, 34-37]. The diverse roles reported for miR-181a in breast cancer progression are summarized in Fig. 1.
Summary of the roles of miR-181a in breast cancer tumor tissue and cell lines. The miR-181a direct targeting genes that have been associated with its anti-oncogenic or oncogenic function are listed with the relevant references. Abbreviations: ATM, ataxia telangiectasia mutated; Bcl-2, B-cell lymphoma 2; BCRP, breast cancer resistance protein; ER+, estrogen receptor–positive; ERK, extracellular-signal regulated kinase; MMP, matrix metalloproteinase; PDCD4, programmed cell death protein 4; PHLDA1, Pleckstrin homology-like domain, family A, member 1; uPA, urokinase-type plasminogen activator
Summary of the diverse roles of miR-181a in breast cancer. Abbreviations: ATM, ataxia telangiectasia mutated; Bcl-2, B-cell lymphoma 2; BCRP, breast cancer resistance protein; MMP-14, matrix metalloproteinase; PDCD4, programmed cell death protein 4; PHLDA1, Pleckstrin Homology-Like Domain, Family A, member 1; uPA, urokinase-type plasminogen activator.
Summary of the diverse roles of miR-181a in breast cancer. Abbreviations: ATM, ataxia telangiectasia mutated; Bcl-2, B-cell lymphoma 2; BCRP, breast cancer resistance protein; MMP-14, matrix metalloproteinase; PDCD4, programmed cell death protein 4; PHLDA1, Pleckstrin Homology-Like Domain, Family A, member 1; uPA, urokinase-type plasminogen activator.
This discrepancy cannot be attributed to different cell types, as different miR-181a functions have been observed in the same cancer cell type. These dissimilar functions may be attributed to the presence of autoregulatory loops [6]. Tsang et al. demonstrated that miRNAs and their targets can form feedback and/or feedforward loops [53]. Therefore, it is plausible that miR-181a is regulated by one or more of its own target genes, such as p53. In fact, the feedforward relationship between miR-181a and p53 has been reviewed in detail by Seoudi et al [6]. Thus, therapeutic strategies employing miR-181a should be developed not only based on the tumor type, but also on the gene expression status and cellular microenvironment in which it is acting. miR-181a can directly target multiple genes and may thus affect multiple pathways and processes concurrently. These targets may also be modulated by a wide range of miRNAs other than miR-181a. Several computational target prediction programs have been developed in recent years to identify miRNA target genes, but their specificity and sensitivity remain low [54]. Although several miR-181a target genes have been identified, the databases remain incomplete due to the high costs of experimental approaches. Understanding miR-181a targets and networks, as well as the expression of miR-181a in different breast cancer subtypes and stages should be the first step in developing therapeutic strategies for breast cancer treatment.
Extracellular miRNA (ex-miRNA) and ex-miR-181a
Extracellular microvesicles (EVs) are membrane surrounded structures released by cells in an evolutionarily conserved manner. They have been characterized by size into three different classes (exosomes: <100 nm; microparticles/microvesicles: <1000 nm; apoptotic bodies: 1–4 μm) [55-57]. EVs can be found in most human biological fluids such as urine, amniotic fluid, malignant ascites, saliva, and blood. Since the discovery of miRNAs within EVs (ex-miRNAs) in 2008, ex-miRNAs have been the subject of much research in the field of molecular and cellular biology. The main reason for this interest is that circulating ex-miRNAs mediate both short- and long-range communication between various cells, and could impact diverse physiological and pathological processes [55, 58-61]. Ex-miRNAs are relatively stable and are present in certain cancers, making them strong candidates as biological markers. So far, the majority of miRNAs detectable in serum and saliva are concentrated in exosomes [62].
Like other ex-miRNAs, ex-miR-181a has received considerable attention in recent years. Thus far, ex-miR-181a has been identified in exosomes derived from breast cancer cells [63], dendritic cells [64], T cells (T cell line J77) [64], stem cells [65, 66], and esophageal cancer cells [67]. It was also found in serum exosomes of patients with ovarian cancer, pancreatic ductal adenocarcinoma, and thyroid cancer [68-70]. ex-miR-181a was found to be one of the twenty most abundant human plasma exosomal miRNAs [71]. Several studies have explored the physiological and pathophysiological roles of ex-miR-181a. For example, ex-miR-181a found in exosomes derived from human adipose mesenchymal stem cells may play a role in inducing apoptosis of cancer cells [65]. In exosomes derived from human bone marrow-derived mesenchymal stem cells, it may serve in the pathogenesis of osteogenic dysfunction [66]. Although efforts have been made to use ex-miR-181a as a biomarker for diagnosing cancer, this potential application is still in its infancy.
We have been working over the past ten years on the pathophysiological roles of lymphocyte-derived microparticles (LMPs), generated from apoptotic human CEM T cells following actinomycin D stimulation [72, 73]. Our investigations have revealed strong anti-angiogenic and anti-neoplastic effects of LMPs [74-79]. We recently performed miRNA sequence analysis and revealed that miR-181a is one of the most abundant ex-miRNAs in LMPs. This is not surprising because miR-181a is highly expressed in the thymus, the primary lymphoid organ where T lymphocyte maturation occurs in the early stages of T-cell differentiation [7, 60, 80-83]. Furthermore, in vitro experiments have shown that overexpression of miR-181a strongly reduces the cell viability of human umbilical vein endothelial cells (HUVEC), human retinoblastoma (Rb) cells, and human breast cancer cells (MCF-7) in a dose-dependent manner. It is interesting to note that, with doses higher than 50 nM, miR-181a completely lost its inhibitory effect; instead it promoted cell growth in variouscancer cell lines in vitro (Fig. 2, unpublished data). This finding is consistent with reports that miR-181a can exhibit different functions within the same cancer cell type.
The inhibitory effect of miR-181a on cell viability. The human breast cancer cell (MCF-7), human retinoblastoma (Rb), and human umbilical vein endothelial cell (HUVEC) were transfected with indicated concentrations of miR-181a mimic or control miRscr for 48 hours; cell viability was assessed using MTT assay. Values were normalized to cell viability of control cells and plotted as mean ± SE. (*p<0.05, ***p<0.01 vs. miRscr).
The inhibitory effect of miR-181a on cell viability. The human breast cancer cell (MCF-7), human retinoblastoma (Rb), and human umbilical vein endothelial cell (HUVEC) were transfected with indicated concentrations of miR-181a mimic or control miRscr for 48 hours; cell viability was assessed using MTT assay. Values were normalized to cell viability of control cells and plotted as mean ± SE. (*p<0.05, ***p<0.01 vs. miRscr).
Altered Serum Levels of miR-181a as a Diagnostic Biomarker
Although direct measurements of tissue biomarkers have greatly improved breast cancer diagnosis, the invasive nature of the diagnostic procedures limits their application (reviewed by Bertoli et al.) [84]. Screening for molecular changes in the patient’s blood is important for early breast cancer detection and prognosis. In fact, miRNAs have been considered as potential biomarkers: miRNAs can be secreted by solid tumors into the surrounding environment; they are stable, retrievable, and can be quantified in fresh serum, archival serum, and plasma samples [85-88].
A growing body of evidence suggests that concentrations of blood-based miRNAs in patients with breast cancer are significantly higher or lower than that in normal individuals, indicating that circulating miRNAs may serve as a novel blood-based biomarker for breast cancer. The significant reduction in miR-181a levels in the blood of patients with breast cancer has been confirmed using different miRNA detection methods by independent studies. Guo et al. compared the relative miR-181a expression levels in serum from healthy controls (n=75) and breast cancer patients (n=152) [89]. Having performed a custom-made stem-loop quantitative PCR after normalization using serum miR-16 levels, they found that miR-181a expression was significantly lower in patients with breast cancer than in healthy controls. In the same study, they also compared the expression levels of miR-181a in serum from breast cancer patients with different clinical TNM classification stages and histological grades; statistical analyses revealed no significant differences. Regarding the miR-181a serum levels and hormone receptor status, there were no significant correlations between miR-181a and ER or PR status in breast cancer. In another study, McDermott et al. detected decreased miR-181a levels by RT-qPCR in the whole blood of 54 patients with luminal A-like breast cancer compared to healthy controls [90]. Ferracin et al. used miRNA absolute quantification with digital PCR technology to analyze the serum miR-181a absolute levels in 60 patients with primary breast cancer, and showed that miR-181a levels were significantly lower compared to healthy controls. This result was further validated in the serum of two independent extended cohorts of patients with breast cancer, one sample was collected in Ferrara (Italy cohort, 2010–2014) and the other in Oklahoma (USA cohort, 2005–2013) [91]. These studies proposed the use of blood-based miR-181a as a diagnostic biomarker for breast cancer. On the contrary, Godfrey et al. prospectively collected serum samples from 205 women who subsequently developed breast cancer and 205 women who remained cancer-free and used microarrays to comprehensively assess known miRNAs. They found miR-181a overexpressed in the samples obtained from breast cancer patients. However, when they attempted to verify this finding in a small independent sample of 5 breast cancer cases and 5 controls, the differences were not statistically significant [92]. Overall, these results suggest that serum miR-181a may represent a novel biomarker for primary breast cancer as well as for early diagnosis. Recent evidence suggests that multiple miRNA-based profiles have better diagnostic and prognostic performance, as well as better sensitivity, than individual miRNA assays [93]. Thus, in combination with other markers, serum miR-181a may improve the sensitivity of breast cancer screening [89].
Although these are promising and exciting results, certain limitations need to be overcome before clinical application. The reproducibility of miR-181a assessments may be affected by: 1) pre-analytical steps such as race, age, and cancer treatments (chemotherapy, surgery, radiation or a combination of these methods) [3, 94]; 2) the handling and technological approach of processing specific samples (mainly serum, plasma) [95, 96]; and 3) methods used for data normalization, i.e. selection of a stable circulating “reference” gene [97, 98]. Indeed, the reference range of miR-181a levels that characterize the healthy state should be determined using a healthy population comparable to subjects demanding a diagnostic procedure. Within the clinical setting the technological approach needs be standardized and more studies need to examine the ‘normal’ level of miR-181a with certain bodily fluids. In addition, the mechanism of miR-181a release into these fluids needs to be characterized.
Conclusion
Breast cancer is a heterogeneous tumor type resulting in a diverse array of malignant features and clinical outcomes. Deciphering the molecular mechanisms that fuel breast cancer development and that act as determinants of aggressiveness is a primary requirement for improving patient management. miR-181a can be detected in extracted tissue samples and bodily fluids, including serum, plasma, and bone marrow [28, 89, 91, 92]. Although the source of the endogenous circulating miR-181a is still unclear, we provide evidence that miR-181a is abundant in LMPs. Since miR-181a may directly target multiple genes concurrently, more investigation is required to determine the miR-181a target genes and networks, which is essential to advance therapeutic strategies.
Abbreviations
ATM (ataxia telangiectasia mutated); BCRP (breast cancer resistance protein); E2 (estradiol); ER (estrogen receptor); ex-miRNA (extracellular miRNA); HER2 (human epidermal growth factor receptor 2); LN (lymph node); LMPs (Lymphocyte-derived MPs); miRNA (microRNA); MMP-14 (Matrix metalloproteinase-14); MS (mammosphere); MX (mitoxantrone); PCR (polymerase chain reaction); PDCD4 (programmed cell death protein 4); PHLDA1 (Pleckstrin Homology-Like Domain, Family A, member 1); PR (progesterone receptor); RT-qPCR (real-time quantitative polymerase chain reaction); TNBC (triple negative breast cancer); uPA (urokinase-type plasminogen activator); UTR (3’-untranslated region).
Acknowledgements
This work was supported by an operating grant to P. Hardy from Canadian Institutes of Health Research (362383).
Disclosure Statement
No potential conflicts of interest were disclosed.