Abstract
Brain receptor patterns for the corticotropin-releasing factor (CRF) receptors, CRF1 and CRF2, are dramatically different between monogamous and promiscuous vole species, and CRF physiologically regulates pair bonding behavior in the monogamous prairie vole. However, it is uncertain whether species differences also exist in the neuroanatomical distribution of the endogenous ligands for the CRF1 and CRF2 receptors, such as CRF and urocortin-1 (Ucn1). We compared the expression of CRF and Ucn1 in four vole species, monogamous prairie and pine voles, and promiscuous meadow and montane voles, using in situ hybridization of CRF and Ucn1 mRNA. Our results reveal that CRF mRNA expression patterns in all four vole species appear highly conserved throughout the brain, including the olfactory bulb, nucleus accumbens, bed nucleus of the stria terminalis, medial preoptic area, central amygdala, hippocampus, posterior thalamus, and cerebellum. Similarly, Ucn1 mRNA primarily localized to the Edinger-Westphal nucleus in all four vole species. Immunocytochemistry in prairie and meadow voles confirmed localization of CRF and Ucn1 protein to these previously identified brain regions. These data demonstrate a striking dichotomy between the extraordinary species diversity of brain receptor patterns when compared to the highly conserved brain distributions of their respective ligands. Our findings generate novel hypotheses regarding the evolutionary mechanisms underlying the neural circuitry of species-typical social behaviors.
Introduction
Microtine rodents provide a useful comparative model to identify the neuroanatomical substrates involved in social behavior. Comparative studies of these species, such as the monogamous prairie (Microtus ochrogaster) and pine (Microtus pinetorum) voles in contrast to their promiscuous sister species, the meadow (Microtus pennsylvanicus) and montane (Microtus montanus) voles, have shed much light on several neurotransmitter systems that modulate social behavior [Insel and Young, 2001; Young and Wang, 2004]. Early research identified neuropeptides such as vasopressin and oxytocin in the regulation of monogamous behaviors, including adult pair bond formation between mates and biparental care of offspring [Insel and Shapiro, 1992; Insel et al., 1994; Young, 1999: Lim et al., 2004a]. A particular pattern of vasopressin V1a receptors (V1aR) has been correlated with monogamy in rodent species, and this is supported by the finding that transgenic mice with a prairie vole-like V1aR distribution also display increased affiliative behavior [Young, 1999; Young et al., 1999]. Moreover, promiscuous vole species with V1aR expression patterns altered to resemble prairie voles display enhanced pair bonding behavior [Lim et al., 2004a]. These data suggest that the specific pattern of V1aR in the brain is responsible for monogamous social behavior in voles.
Recently it has been shown that a third neuropeptide system, corticotropin-releasing factor (CRF), also regulates social attachment in voles. CRF and related neuropeptides, Urocortin-1 (Ucn1), Urocortin-2 and Urocortin-3 interact with two main classes of receptors in the brain, CRF1 and CRF2 [for recent reviews see Bale and Vale, 2004; Gysling et al., 2004]. CRF and Ucn1 have been extensively studied in relation to stress, food consumption, fear, and learning and memory paradigms, but much less is known about their roles in social behavior [Spina et al., 1996; Radulovic et al., 1999; Benoit et al., 2000; D’Anna et al., 2005; Weitemier and Ryabinin, 2006]. A recent experiment showed that intracerebroventricular (i.c.v.) administration of CRF facilitates partner preference in male prairie voles [DeVries et al., 2002]. In agreement with the role of CRF systems in pair bonding, we have recently identified dramatic species differences in the distribution of CRF1 and CRF2 receptors between two promiscuous and two monogamous species of voles [Lim et al., 2005a]. The most significant species differences in CRF1 and CRF2 were in the nucleus accumbens (NAcc), where both promiscuous species had dense CRF1 binding, whereas both monogamous species had dense CRF2 binding [Lim, pers. observ.]. Subsequent pharmacological manipulation of CRF1 and CRF2 in the NAcc has confirmed the necessity of accumbal CRF2 for partner preference in monogamous species [Lim, pers. observ.].
These findings suggest that, analogous to V1aR brain patterns, a ‘monogamous-typical’ CRF1 and CRF2 receptor distribution could also have evolved in monogamous vole species. This is compelling in light of previous studies implying that species-typical patterns of vasopressin and oxytocin receptors within the brain are correlated with, if not the cause of, monogamous social organization [Insel and Shapiro, 1992; Insel et al., 1994; Young et al., 1999; Lim et al., 2004c]. However, in contrast to receptor distribution, the distribution of endogenous vasopressin and oxytocin ligands appear quite conserved across species, suggesting that differences in peptide action in a given brain region would be determined almost exclusively by the presence or absence of their respective receptors [Wang et al., 1996].
We sought to determine if this same comparative phenomenon also applied to the CRF system, using in situ hybridization and immunohistochemistry for CRF and Ucn1 in the brain of several vole species. Recent studies have shown that, unlike vasopressin and oxytocin, the distribution of cells expressing CRF-like peptides and their fibers shows some degree of variability not only from species to species, but even among different genetic strains within species [Bachtell et al., 2002; Weitemier et al., 2005]. Therefore, we hypothesized that the levels of CRF and CRF-related peptides in brain regions potentially involved in regulation of social behaviors would differ between monogamous and promiscuous species of voles. On the other hand, if we observed high conservation of CRF and Ucn1 distribution across voles, then this would further highlight variation in receptor distribution as the primary mechanism for driving diversity in social behavior.
Materials and Methods
Subjects
The animals were adult male and female prairie voles (70–100 days of age) from our laboratory breeding colony that were descendents of field-captured voles from Illinois, USA. Adult male and female meadow voles of the same age were derived from stock originally obtained from the laboratory breeding colony of Dr. Zuoxin Wang at Florida State University. Adult male and female pine vole brains were obtained from the laboratory breeding colony of Dr. John Vandenbergh at North Carolina State University. Montane vole brains were taken from a terminated laboratory colony originally derived from stock maintained by the National Institutes of Health. After weaning at 21 days of age, subjects were housed in same-sex sibling pairs or trios with water and Purina rabbit chow provided ad libitum. All cages were maintained on a 14:10 light:dark cycle with the temperature at 20°C. Sixteen adult animals were used in the generation of data for the in situ hybridization experiments (n = 4 for each of the four vole species, with 2 males and 2 females per species). Additionally, a total of 22 adult animals were used for the immunohistochemistry experiments (n = 5 female prairies, n = 6 male prairies, n = 5 female meadows, n = 6 male meadows). All animals were sacrificed between the hours of 10:00 and 14:00 at basal conditions. Adult pine and montane vole brains were not in an adequately perfused condition and therefore could not be included in the immunohistochemistry study. We were unable to process any vole tissue adequately for Urocortin-2 or Urocortin-3 in situ hybridization or immunocytochemistry and therefore, did not include these data in the study. All experiments were approved by the institutional guidelines set by the animal care and use committee of Emory University and conformed to the guidelines set by the NIH.
CRF Riboprobe in situ Hybridization
Adult male and female voles (n = 16, four of each species) were rapidly decapitated and their brains removed and snap-frozen on powdered dry ice. Brains were sectioned from olfactory bulbs through the cerebellum at 20 µm on a cryostat at –20°C and mounted on Fisher superfrost plus microscope slides. The full-length clone for the rat CRF gene was constructed from a 1.2 kb Eco RI fragment of a full-length rat CRF cDNA subcloned into a pBluescript II-SK+ plasmid (obtained as a gift from W. Vale, Salk Institute, La Jolla, Calif.). In situ hybridization was performed with an antisense riboprobe as previously described [Ressler et al., 2002]. Briefly, an 35S-UTP-labeled riboprobe was prepared from the linearized clone using SP6 polymerase at high specific activity by using only radioactive UTP in the polymerase reaction. The RNA probe was base-hydrolyzed to average lengths of 50–100 bp and isolated using a Sephadex gravity flow column. Hybridizations were incubated under parafilm at 55°C overnight. Slides were then stringently washed, dried, and placed against Kodak MR autoradiography film for 3 days.
Ucn1 Oligoprobe in situ Hybridization
One adjacent set of slides was processed for urocortin-1 (Ucn1) in situ hybridization. The protocol used was similar to that previously described for other oligoprobes [Lim et al., 2004b]. Slides were pre-treated by fixing in 4% paraformaldehyde (5 min), and then rinsed for 5 min in 0.1 M phosphate buffered saline (1× PBS, pH 7.4 ), treated for 10 min with 0.1 M triethanolamine plus 0.25% acetic anhydride, and rinsed in 0.03 M Na citrate in 0.3 M NaCl (2× SSC). Slides were then dehydrated in ascending concentrations of ethanol, lipids were removed by chloroform and air-dried. Slides were then pre-hybridized with parafilm covering 150 µl of a solution containing 50% formamide, 4× SSC, 1× Denhardt’s solution, 10 mg/ml yeast transfer RNA, 10% dextran sulfate, 0.3 M NaCl, 10 mM Tris, and 10 mM dithiothreitol (DTT) for 1 h at 37°C. Following pre-hybridization, slides were washed twice in 2× SSC, dehydrated in ethanol, and air-dried. The hybridization solution consisted of the same components as the pre-hybridization solution, except with the substitution of 2 mg/ml yeast transfer RNA and the addition of 10 pmol/ml concentration of oligodeoxyribonucleotide probe for the Ucn1 gene. Each slide was treated with 200 µl of hybridization solution covered with parafilm overnight at 37°C. The Ucn1 probe consisted of 48 bases (AAA TAA CCA CTC AGA GTA TTC AGG GTG ACT CTG GGC GCG CTG CAC CGC) and was complementary to the rat mRNA encoding base pairs 571–618 of the Ucn1 message (Genbank accession number NM021290). The probe was labeled at the 3′ end with 35S-dATP (NEN/PerkinElmer, MA) using terminal deoxynucleotidyl transferase (Promega, WI). After 4 × 15 min rinses in 1× SSC at 50°C and a spinning wash at room temperature, dehydration in ascending ethanols, and air-drying, slides were apposed to Kodak BioMax MR film for 2 days to generate autoradiograms.
Semi-Quantitative Analysis
Autoradiograms were blinded to the experimenter and quantified on a scale of 0–4 based on relative densitometry of decompositions per minute of radioactivity. The following ten brain regions were semi-quantified for CRF mRNA density in each animal: olfactory bulb, nucleus accumbens, bed nucleus of the stria terminalis, medial preoptic nucleus, paraventricular nucleus of the hypothalamus, hippocampus, central nucleus of the amygdala, posterior thalamus, median raphe, and cerebellum. One brain region, the Edinger-Westphal nucleus, was semi-quantified for Ucn1 mRNA. No gross sex differences were observed, and therefore both sexes were averaged into a single semi-quantitative value for each brain region of the four different species. Gross distribution and overall patterns of brain peptide were assessed, and further emulsion microanalysis was not performed.
Photomicrograph Production Details
Digital images were obtained from film autoradiograms using a B-95 Northern Light light box (Imaging Research, Inc., Ontario, Canada) and a SPOT camera (Diagnostic Instruments, Inc., Michigan, USA) connected to a computer through the RT SPOT power supply. Digital images obtained from microscope slides were similarly taken using the SPOT camera setup. Images were then imported into Adobe Photoshop 7.0 (Adobe Systems Inc., California, USA), cropped to the correct size, and minimally adjusted for brightness and contrast in order to clarify the scientific points of interest. All figure text, arrows, and scale bars were added using Adobe Illustrator 10.0 (Adobe Systems Inc., California, USA).
CRF and Ucn1 Immunohistochemistry
Adult prairie and meadow voles were deeply anesthetized between the hours of 10 am and 2 pm with isoflurane and perfused with saline followed by 2% paraformaldehyde in 10 mM (pH 7.4) phosphate-buffered saline (PBS). Dissected brains were postfixed overnight in the 2% paraformaldehyde/PBS solution and cryoprotected in 30% sucrose/PBS. 30-µm free-floating coronal sections were sliced on a cryostat and processed for immunohistochemistry according to standard protocols [Ryabinin et al., 1997; Weitemier et al., 2005] with adjustments for vole tissue made in preliminary experiments. Briefly, endogenous peroxidase activity was quenched by 15-min incubation with 0.3% hydrogen peroxide. For the Ucn1-specific antibody, blocking was performed by a five-hour incubation with 2% Bovine Serum Albumin, 0.1% heparin, 0.01% Triton X-100 in PBS. CRF-specific antibody blocking was performed by a five-hour incubation with 4.5% goat serum, 0.3% Triton X-100 in PBS. The primary rabbit antibodies recognizing Ucn1 (Sigma-Aldrich, St. Louis, Miss., USA) were diluted to 1:5,000. The primary antibodies recognizing CRF (Peninsula Laboratories, San Carlos, Calif., USA) were used in the dilution of 1:15,000. Biotinylated anti-rabbit secondary antibodies (Vector Laboratory Inc., Burlingame, Calif., USA) were used to detect the primary antibodies. Detection of the secondary antibodies was made using Vectastain ABC kit (Vector), and enzymatic development was done with the Metal Enhanced DAB kit (Pierce, Rockford, Ill., USA). The specificity of immunostaining was assessed by a complete lack of immunoreactivity in areas not known to express CRF or Ucn1. Futhermore, control preabsorption experiments for these antibodies were previously performed [Bachtell et al., 2003].
CRF2 Receptor Autoradiography
An adjacent set of fresh-frozen slides was processed for CRF2 receptor autoradiography. CRF2 receptor binding was measured by incubating 125I-Tyr⁰-sauvagine, which binds to both CRF1 and CRF2 receptors, with unlabeled CP-154,526-1 (butyl-[2,5-dimethyl-7-(2,4,6-trimethylphenyl)-7H-pyrrolo[2,3-d]pyrimidin-4-yl]-ethylamine), a selective CRF1 receptor antagonist which was synthesized at Emory University and kindly provided by Michael J. Owens, Ph.D. This protocol has been successfully employed in vole brains [Lim et al., 2005]. Briefly, slides were lightly fixed in 0.1% paraformaldehyde-PBS (pH 7.4), rinsed twice in 50 mM Tris base (pH 7.4), and then incubated in tracer for 2 h. The tracer buffer consisted of a 50 mM Tris base, 10 mM MgCl, 0.1% bovine serum albumin, 0.05% bacitracin, plus 0.2 nM125I-Tyr⁰-sauvagine (PerkinElmer/NEN, Boston, Mass., USA) and 1 µM CP-154,526. Slides were then rinsed with 50 mM Tris base plus 10 mM MgCl (pH 7.4), dipped in deionized H20, blown dry with cool air, and apposed to Kodak MR film for 85 h with 125I microscale standards (PerkinElmer/NEN, Boston, Mass., USA).
Results
CRF Riboprobe in situ Hybridization
CRF in situ hybridization throughout the brain revealed that the distribution pattern of CRF mRNA across all four vole species was highly conserved. All ten brain regions analyzed in all species expressed CRF mRNA. The results of a semi-quantitative analysis of mRNA signal in several brain regions are presented in table 1. Representative brain sections throughout the brain for all four vole species are shown in figures 1 and 2.
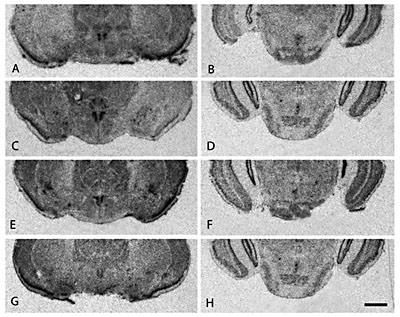
CRF in situ hybridization in the nucleus accumbens (A–D), bed nucleus of the stria terminalis/medial preoptic area (E–H), central nucleus of the amygdala (I–L), and posterior nuclei of the thalamus (M–P) in four vole species. Note the similarity in brain distribution of CRF mRNA in the various brain regions among the prairie voles (A, E, I, M), pine voles (B, F, J, N), meadow voles (C, G, K, O), and montane voles (D, H, L, P). Scale bar = 1 mm.
CRF in situ hybridization in the nucleus accumbens (A–D), bed nucleus of the stria terminalis/medial preoptic area (E–H), central nucleus of the amygdala (I–L), and posterior nuclei of the thalamus (M–P) in four vole species. Note the similarity in brain distribution of CRF mRNA in the various brain regions among the prairie voles (A, E, I, M), pine voles (B, F, J, N), meadow voles (C, G, K, O), and montane voles (D, H, L, P). Scale bar = 1 mm.
CRF in situ hybridization in the paraventricular nucleus of the hypothalamus (A, C, E, G) and median raphe nucleus (B, D, F, H) in four vole species. Note the similarity in brain distribution of CRF mRNA in both brain regions among the prairie voles (A, B), pine voles (C, D), meadow voles (E, F), and montane voles (G, H). Scale bar = 1 mm.
CRF in situ hybridization in the paraventricular nucleus of the hypothalamus (A, C, E, G) and median raphe nucleus (B, D, F, H) in four vole species. Note the similarity in brain distribution of CRF mRNA in both brain regions among the prairie voles (A, B), pine voles (C, D), meadow voles (E, F), and montane voles (G, H). Scale bar = 1 mm.
Most rostrally, the olfactory bulbs highly expressed CRF mRNA in what appeared to be the granular and plexiform layers in prairie, pine, meadow, and montane voles (data not shown). CRF mRNA signal was detected in the ventral forebrain in all four species in areas corresponding to the nucleus accumbens (NAcc) septal pole, caudal ventral pallidum, and medial bed nucleus of the stria terminalis (BnST) (fig. 1A, B, C, D). No overt sex or species differences in ligand expression were observed. Medial BnST and rostral medial preoptic area (MPOA) showed moderate CRF mRNA signal, with no noted species or sex differences in the degree of CRF mRNA expression (fig. 1E, F, G, H). The paraventricular nucleus of the hypothalamus (PVN) also showed dense CRF mRNA signal, with monogamous prairie and pine voles appearing to express slightly more CRF mRNA in the PVN than their promiscuous counterparts (fig. 2A, C, E, G). CRF mRNA signal was detected in all hippocampal fields including CA1, CA2, CA3, and dentate gyrus as well as the central nucleus of the amygdala (CeA) (fig. 1I, J, K, L). Hybridization signal was detected in the posterior nuclei of the thalamus, including subdivisions of the lateral geniculate nucleus with slightly varying degrees among the four vole species (fig. 1M, N, O, P). In the hindbrain, CRF in situ hybridization signal was present in the median raphe nucleus (MR) in all four species (fig. 2B, D, F, H). However, monogamous prairie and pine voles appeared to show greater CRF mRNA signal in the MR than promiscuous meadow and montane voles. CRF mRNA was also highly expressed in the granular layers of the cerebellum in all four vole species (data not shown).
There were also several brain regions, including the prefrontal and cingulate cortices, substantia nigra, locus coeruleus, and periaqueductal grey that did not show CRF mRNA signal in voles although previously reported to show signal in other rodent species [Swanson et al., 1983; Merchenthaler, 1984; Sakanaka et al., 1987; Morin et al., 1999; Steckler and Holsboer, 1999].
Ucn1 Oligoprobe in situ Hybridization
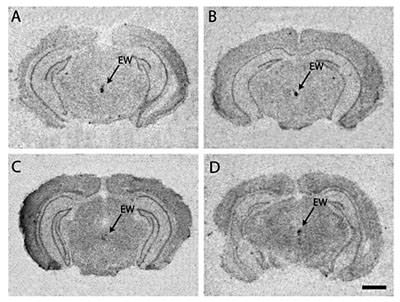
Ucn1 in situ hybridization was detected almost exclusively in the Edinger-Westphal nucleus (EW) of the brain in all four vole species (fig. 3). Prairie, pine, and montane voles appeared to have somewhat denser Ucn1 mRNA signal in the EW than meadow voles. Some Ucn1 mRNA signal was detected at lower levels throughout the hippocampus, including the CA1 subfield, but was not found in any other brain region (data not shown). No sex differences in Ucn1 mRNA signal were observed.
Urocortin-1 in situ hybridization in the Edinger-Westphal nucleus in the monogamous prairie vole (A), pine vole (B), and promiscuous meadow vole (C) and montane vole (D). Note the similarity in brain distribution of Ucn1 mRNA among the four vole species. Scale bar = 1 mm.
Urocortin-1 in situ hybridization in the Edinger-Westphal nucleus in the monogamous prairie vole (A), pine vole (B), and promiscuous meadow vole (C) and montane vole (D). Note the similarity in brain distribution of Ucn1 mRNA among the four vole species. Scale bar = 1 mm.
There were several brain regions that did not show Ucn1 mRNA signal in voles that have been previously reported in other rodent species after colchicine induction. These regions include the lateral septum, medial amygdala, substantia nigra, and the cerebellum (table 1) [Kozicz et al., 1998; Bittencourt et al., 1999; Morin et al., 1999].
CRF Immunohistochemistry
CRF immunocytochemistry confirmed the general pattern of brain distribution revealed by CRF in situ hybridization. CRF-immunoreactive cells were identified in the PVN, a brain region known to express this peptide in other rodent species, with a greater cell density in prairie voles compared to meadow voles (fig. 4A, B). Confirming the mRNA findings, prairie voles exhibited a high level of immunoreactivity in the median raphe (fig. 4C) compared to meadow voles (fig. 4D). Strong CRF-immunoreactive staining was identified in the BnST (fig. 4E, F), CeA (fig. 4G, H), with no overt species differences observed. Consistent with reports from other rodents, cellular immunoreactivity in BnST and CeA was likely obscured by high background from fiber staining [Cummings et al., 1983; Sawchenko and Swanson, 1985; Asan et al., 2005; Hayes et al., 2005]. CRF-immunoreactive fiber staining was also identified within the septal pole of the nucleus accumbens in both prairie and meadow voles (data not shown). Weak CRF-positive staining was found in what appeared to be the microcellular tegmental area, neocortical areas, and thalamic areas (data not shown). Staining was noted in both prairie and meadow voles and in both sexes.
CRF-immunoreactive neurons in the paraventricular nucleus of hypothalamus (A, B), median raphe (C, D), bed nucleus of the stria terminalis (E, F), and central nucleus of the amygdala (G, H) in prairie (A, C, E, G) and meadow (B, D, F, H) voles. Scale bar = 200 µm.
CRF-immunoreactive neurons in the paraventricular nucleus of hypothalamus (A, B), median raphe (C, D), bed nucleus of the stria terminalis (E, F), and central nucleus of the amygdala (G, H) in prairie (A, C, E, G) and meadow (B, D, F, H) voles. Scale bar = 200 µm.
Ucn1 Immunohistochemistry
Brain distribution of Ucn1 mRNA signal was consistent with Ucn1 peptide immunoreactivity in that both mRNA and peptide were detected almost exclusively in the EW nucleus in all four species and both sexes. Confirming the mRNA findings, prairie voles exhibited a somewhat higher level of Ucn1 immunoreactivity in the EW nucleus compared with meadow voles, a difference perhaps attributable to an increase in the number of Ucn1-expressing neurons (fig. 5A, B).
Urocortin-1 expression in the Edinger-Westphal nucleus in prairie (A) and meadow (B) voles. Scale bar = 200 µm.
Urocortin-1 expression in the Edinger-Westphal nucleus in prairie (A) and meadow (B) voles. Scale bar = 200 µm.
CRF2 Receptor Binding in NAcc
Given previous data implicating CRF2 receptors in the nucleus accumbens (NAcc) in the regulation of partner preference by monogamous prairie voles, we processed adjacent brain sections for CRF2 receptor autoradiography to compare the location of CRF mRNA with CRF2 receptor binding (fig. 6). CRF mRNA in the NAcc was located approximately 1 mm caudal to the region of dense CRF2 receptor binding in the two monogamous vole species (fig. 6). CRF2 receptor binding was absent throughout the NAcc in promiscuous voles, despite dense CRF mRNA signal in the caudal NAcc (data not shown). Thus, it appears that the major differences in CRF patterns between monogamous and promiscuous voles lie within the distribution pattern of receptors, in contrast to relative conservation of their respective ligands.
Paired sections stained for CRF in situ hybridization (A, C), and CRF2 receptor autoradiography (B, D) at the level of the nucleus accumbens in monogamous prairie (A, B), and pine (C, D) voles. Note the presence of CRF mRNA signal in the same vicinity of CRF2 receptor binding. Promiscuous meadow and montane voles lack CRF2 binding in the nucleus accumbens. Scale bar = 1 mm.
Paired sections stained for CRF in situ hybridization (A, C), and CRF2 receptor autoradiography (B, D) at the level of the nucleus accumbens in monogamous prairie (A, B), and pine (C, D) voles. Note the presence of CRF mRNA signal in the same vicinity of CRF2 receptor binding. Promiscuous meadow and montane voles lack CRF2 binding in the nucleus accumbens. Scale bar = 1 mm.
Discussion
Our report describes for the first time the distribution of CRF and Ucn1 in the vole brain. Patterns of peptide appeared to be highly conserved across four vole species, with all 10 CRF-positive brain regions, and one Ucn1-positive brain region, showing some degree of expression in all species. Likewise, brain regions reported to express CRF or Ucn1 in other rodent species uniformly did not show detectable signal in any of the four vole species.
For the most part, vole species expressed CRF and Ucn1 within the major brain regions identified in rat and mouse brain, such as the olfactory bulb, BnST, hippocampus, CeA, PVN, raphe, cerebellum, and Edinger-Westphal nucleus [Swanson et al., 1983; Merchenthaler, 1984; Wong et al., 1996; Morin et al., 1999; Allen, 2006]. Voles also highly expressed CRF within the medial preoptic area, an area not previously reported to express CRF in other rodents. Conversely, there were several brain regions such as the prefrontal cortex, cingulate cortex, locus coeruleus, substantia nigra, and periaqueductal grey in which rats were reported to express CRF, whereas our voles did not [Swanson et al., 1983; Merchenthaler, 1984; Wong et al., 1996; Morin et al., 1999; Allen, 2006]. Within the rat brain, Ucn1 mRNA has been reported within the medial and lateral septum, PVN, amygdala, substantia nigra, superior colliculus, red nucleus, and cerebellum, although these studies used colchicine to enhance cellular expression [Wong et al., 1996; Yamamoto et al., 1998; Kozicz et al., 1998; Morin et al., 1999; Bittencourt et al., 1999]. The lack of detection of these regions in vole brain could be due to a variety of factors, including a high signal to noise ratio on the autoradiograms, lack of sequence specificity of the hybridization probe, or in fact, true species differences between voles and rats. In any case, however, it is notable that all four vole species are consistent in their lack of expression compared to rat.
Within voles, a few brain regions did show some variation in the degree to which CRF and Ucn1 were expressed, such as within the MR and EW nucleus. It is conceivable that these more subtle species differences could contribute to interspecies variation in social behavior. Monogamous and promiscuous voles differed in the degree to which CRF was expressed in the MR, in both in situ hybridization and immunocytochemistry. Interestingly, high levels of CRF peptide immunoreactivity in MR of prairie voles were detected by us, but have not been seen in other species although some studies have reported lower numbers of CRF-positive cells in rat MR [Sakanaka et al., 1987]. In this regard, the lower levels of CRF expression in MR of meadow and montane voles is more congruent with that observed in other species. The neurons within MR send their projections to several areas, including the diagonal band of Broca, lateral habenula, lateral hypothalamus, mammillary nucleus of hypothalamus, centrolateral nucleus of thalamus, interpeduncular nucleus and the ventral tegmental area [Morin et al., 1999; Vertes et al., 1999]. The specific targets of CRF-positive neurons within MR in prairie voles and their potential contribution to social behavior bear further investigation.
Likewise, subtle species differences in Ucn1 immunoreactivity within EW could also contribute to species differences in social behavior. Major projection targets of EW include the lateral septum and dorsal raphe, both of which are heavily involved in the regulation of social behaviors, including pair bonding in voles [Czlonkowski et al., 1975; File and Deakin, 1980; Albert and Walsh, 1984; Bittencourt et al., 1999; Bachtell et al., 2004; Weitemier and Ryabinin, 2006]. Blockade of vasopressin receptors in the lateral septum prevents partner preference formation in prairie voles [Liu et al., 2001]. Given the major Ucn1 projection from EW to the lateral septum, it appears that Ucn1 and vasopressin systems could be neuroanatomically linked and therefore potentially interact during social behavior. Thus, these brain regions are attractive potential candidates for identifying relevant circuits involved in CRF-regulated pair bond formation and other involved neuromodulators. However, despite these few species differences in CRF and Ucn1 within the PVN, MR, and EW, more striking is the extraordinary conservation of ligand expression throughout the rest of the brain, especially in comparison to the dramatic variation observed in CRF receptor distribution [Lim et al., 2005].
What about brain regions known to be involved in pair bond formation? CRF mRNA was detected in the nucleus accumbens at similar levels in all four vole species. The site of CRF mRNA expression was found in close proximity to a dense region of CRF2 receptors expressed in monogamous prairie and pine voles, but lacking in promiscuous meadow and montane voles [Lim et al., 2005]. Microinjections of CRF directly into NAcc in monogamous prairie voles, presumably stimulating the dense patch of CRF2 receptors, accelerate pair bond formation [Lim, pers. observ.]. The presence of CRF mRNA and CRF fiber staining in NAcc suggests that endogenous synthesis and release of CRF within this region can potentially activate CRF2 receptors. Thus, although all four vole species express CRF ligand within NAcc to a similar degree, the presence of dense CRF2 receptors in only the monogamous voles would activate different neural circuits during CRF release, thereby allowing for the control of different behaviors.
Our results demonstrate definitively that there are few species differences in CRF-like peptide distribution at baseline conditions; however, it is currently unknown whether species differences could emerge following exposure to social stimuli, such as before or after a pair bonding event. Indeed, preliminary studies in pair-bonded male prairie voles have shown that CRF mRNA levels in NAcc change when the animal is separated from its partner [Bosch et al., 2005]. Future studies are currently underway to examine these questions.
Why are CRF and Ucn1 so highly conserved, whereas their receptor subtypes CRF1 and CRF2 are so dramatically different among species? Although it remains to be demonstrated whether other CRF-like peptides such as Ucn2 and Ucn3 are also highly conserved, this phenomenon has been observed with other neuropeptide systems, including oxytocin and vasopressin. In the case of vasopressin and its V1aR, vasopressin peptide distribution is conserved within vole species and across other rodents; in contrast, V1aR distribution is so varied that no two species have the exact same distribution [Insel and Shapiro, 1992; Insel et al., 1994; Wang et al., 1996; Goodson and Bass, 2001]. Recent studies manipulating the vole V1aR distribution have demonstrated that the species-specific pattern of V1aR determines affiliative or partner preference behavior [Young et al., 1999; Lim et al., 2004c], and even within a species, individual variation in V1aR distribution appears correlated with aspects of social behavior [Hammock and Young, 2005; Hammock et al., 2005].
Variation in V1aR distribution has been linked to variation in V1aR gene sequence. Monogamous prairie and pine vole V1aR gene sequence contains a microsatellite region located in the 5′ regulatory flanking sequence that is abbreviated in promiscuous meadow and montane voles [Young et al., 1999]. Intraspecific variation in the length of this microsatellite region in prairie voles has been correlated with particular V1aR brain patterns and variability in social behavior [Hammock and Young, 2005; Hammock et al., 2005]. The insertion of microsatellite sequences within gene promoters could enable or prevent the binding of different transcription factors, and in turn regulate gene expression differentially across brain regions and neural circuits. It is possible that such unstable microsatellites might exist within other neuropeptide receptor genes, although further studies will be needed to confirm this hypothesis. We would also predict that, in contrast, regulatory gene sequences for neuropeptide ligands would be highly conserved across species.
Why variability in receptor patterns, and not ligand patterns? Receptor patterns could be more easily altered resulting in differences in social behavior, particularly because frequently more than one receptor subtype might exist for any given ligand. In contrast, dramatic changes in peptide patterns could potentially jeopardize several major neural circuits, as well as peripheral functions. For example, a large change in AVP release patterns could potentially affect peripheral V1aR regulated blood pressure and V2 receptors in the kidney, which might have dramatic physiological or deleterious effects on the organism. Manipulating one receptor subtype would allow the same ligand pattern to preserve its original function in other receptor subtypes, while conferring variation in only one or a few neurocircuits at a time.
It is interesting that this phenomenon of variation in neuropeptide receptors has not been observed for other small molecule neurotransmitter systems in the brain, such as the monoamines or catecholamines. In vole species, neuroanatomical mapping has been performed for benzodiazepine receptors (GABA), dopamine receptors, dopamine transporter, serotonin transporter, and acetylcholinesterase activity, and no species differences exist in the gross distribution of such systems [Insel and Shapiro, 1992; Lim and Young, pers. observ.]. This is consistent with the hypothesis that small molecule neurotransmitter systems are evolutionarily ancient and control more fundamental processes such as motor reflexes, respiration, and excitation and inhibition of neural circuits; there is much less variation in these general physiological processes across taxa due to natural selection for efficiency in these behaviors.
In contrast, oxytocin, vasopressin, and CRF receptors regulate species-typical social behaviors that vary widely across taxa. The particular repertoire of social behaviors such as courtship behavior, territorial aggression, mating and parental care can be unique to each species of animal as a result of any number combinations of receptor patterns in the brain. Wide variation can exist in these behaviors without major effects on fundamental physiology or proximate functions, but instead can have enormous impact upon the ultimate fitness of the species. Depending on the factors influencing natural selection and sexual selection, variation in neuropeptide receptor distribution could provide a mechanism for the rapid evolution of unique social behaviors and even speciation.
Acknowledgments
The authors would like to thank Drs. Stephen Harvey and John G. Vandenbergh at North Carolina State University for their generous donation of pine vole tissue. From Emory University, we would like to thank Dr. Michael J. Owens for graciously providing us with the CP-154,526 compound, and Dr. Wylie Vale from the Salk Institute for the gift of the CRF plasmid. Supported by NIH MH65050 to MML, AA013738 to AER, NSF STC IBN-9876754 and Yerkes Center Grant RR00165.