Abstract
Gene discoveries in cancer have the potential for clinical and public health applications. To take advantage of such discoveries, a translational research agenda is needed to take discoveries from the bench to population health impact. To assess the current status of translational research in cancer genetics, we analyzed the extramural grant portfolio of the National Cancer Institute (NCI) from Fiscal Year 2007, as well as the cancer genetic research articles published in 2007. We classified both funded grants and publications as follows: T0 as discovery research; T1 as research to develop a candidate health application (e.g., test or therapy); T2 as research that evaluates a candidate application and develops evidence-based recommendations; T3 as research that assesses how to integrate an evidence-based recommendation into cancer care and prevention; and T4 as research that assesses health outcomes and population impact. We found that 1.8% of the grant portfolio and 0.6% of the published literature was T2 research or beyond. In addition to discovery research in cancer genetics, a translational research infrastructure is urgently needed to methodically evaluate and translate gene discoveries for cancer care and prevention.
Introduction
Gene discoveries in germ line and tumor cells are rapidly being reported for a wide variety of human cancers, both common and rare [1,2,3,4]. In addition, we are experiencing a rapid proliferation of research in areas of cancer gene expression [5,6], epigenomics [7] and proteomics [8]. These discoveries, if validated, carry great promise for clinical and public health applications in the form of improved risk prediction, early detection, more precise diagnosis and prognosis estimation, and targeted therapeutics [9]. To reap any health benefit from genomic discoveries for cancer treatment and prevention, discovery research must be accompanied by carefully and methodically conducted translational research, to move research from the bench to the bedside, and to the population at large [10]. The Institute of Medicine (IOM) and other advisory groups have become increasingly concerned about the chasm between basic science discoveries and translation to clinical medicine and population health [11]. In this report, we evaluated the current status of translational research in cancer genetics. We present an analysis of the National Cancer Institute (NCI) grant portfolio as well as the cancer genetic research articles published in the peer-reviewed literature. For the purpose of this report, we use the term translational research; however, we consider it interchangeable with translation research as described by Khoury et al. [12] and elsewhere.
Methods
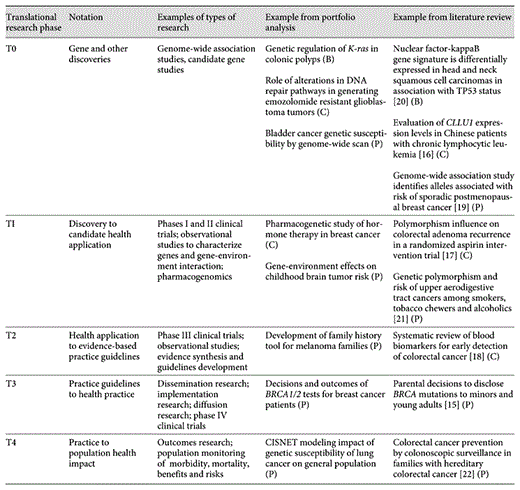
We classified the cancer genetic research into a continuum of categories from discovery research (denoted here as T0) through 4 phases of genomics translational research (denoted T1 through T4) as described by Khoury et al. [12]. This translational research pathway was adapted from previous publications describing general phases of translational research from basic science to population health [12,13,14]. We applied this schema to the existing NCI grant portfolio and cancer literature in 2007, the most recent year with complete data for our analysis. The continuum of translational research is shown in figure 1. Here we present the cancer continuum as a circular diagram to reflect that translation is not a linear process and that stages of translation can provide feedback loops to one another. Specific examples of translational research for cancer genetics outlined in table 1[15,16,17,18,19,20,21,22] are based on our portfolio analysis and literature review.
The continuum of translational research in cancer genetics: types of research and examples from our portfolio analysis and literature review (B = basic; C = clinical; P = population)
The continuum of genetics translational research from gene discovery to reducing the burden of disease in a population.
The continuum of genetics translational research from gene discovery to reducing the burden of disease in a population.
The 5 research categories that we employed are defined as follows. T0 comprises gene discovery such as genome-wide association studies or candidate gene studies. T1 research includes studies that attempt to take discoveries into developing candidate application that can be utilized in cancer care and prevention, including the development of diagnostics (e.g., screening or diagnostic tests) or interventions (e.g., drugs and other interventions). These studies include biological, clinical and epidemiological studies to validate and characterize discoveries in clinical and population settings. They also include phase I and II clinical trials for the development of therapeutic agents. T2 research includes the evaluation of candidate applications (such as predictive or diagnostic tests) for their validity and utility leading to the development of evidence-based recommendations. T2 research includes observational studies, evidence synthesis and as well as phase III clinical trials. T3 research includes studies that demonstrate integration of evidence-based recommendations into the practice of cancer care and prevention (e.g., dissemination, implementation or diffusion research). T4 research evaluates whether a clinical application leads to improved patient outcomes in clinical practice or overall reduction in morbidity and mortality at the population level (e.g., outcomes research, cost-effectiveness, surveillance and phase IV clinical trials). Additional details are available in Khoury et al. [12].
NCI Portfolio Analysis
We searched for potentially relevant grants from 3 internal data sources: the NCI’s Division of Extramural Activities’ Research Analysis and Evaluation Branch (REAB) data; the NCI Portfolio Management Application (PMA) 13.3; and the NIH Query/View/Report (QVR) system (http://qvr.cit.nih.gov/qvrhelpdocs/index.htm). Results from each database were combined and duplicates were removed. To be included in our analysis, the grants had to be funded by NCI; be cancer related; awarded in Fiscal Year 2007; be a research or project grant (i.e., R, U or P mechanisms); and involve human subjects. Additionally the title or abstract had to include one of the following keywords: biomarker, gene, genetic, genome, genomic, epigenetic, epigenomic, gene mapping, genetic testing or personalized health. Grants were deemed irrelevant to our analysis if there was no human component; were not cancer focused; or the grant was administrative in nature. Abstracts for each grant were read and coded into translational research phase (T0/T1/T2/T3/T4) and study type (basic/clinical/population). Four individuals each reviewed a quarter of all grant abstracts (n = 1,844) for inclusion into the portfolio analysis and the fifth member of our team reviewed a random 10% sample of all grants for quality control. Reviewers categorized each relevant abstract (n = 1,019) by translational research phase and study type. Abstracts categorized as T2 and beyond were further categorized as being either therapeutic or diagnostic. All 5 reviewers convened to refine variable definitions and resolve discrepancies.
Literature Review
We searched for potentially relevant abstracts from PubMed (www.ncbi.nlm.nih.gov/pubmed/) and the HuGE Literature Finder (www.hugenavigator.net/HuGENavigator/startPagePubLit.do). To be included in our analysis, publications had to be cancer related; published in 2007; and involve human subjects. Additionally, the title or abstract had to include one of the following keywords: biomarker, gene, genetic, genome, genomic, epigenetic, epigenomic, gene mapping, genetic testing and personalized health.
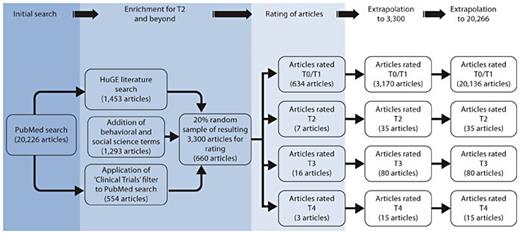
Our PubMed search based on the above keywords for cancer-related, human subjects research in 2007 resulted in over 20,266 articles focusing on cancer genetics published in 2007. We further attempted to find all T2 research and beyond by enriching our search for T2–T4 abstracts using the following 3 steps. First, we applied the ‘Clinical Trials’ filter in PubMed, which selects for clinical trials, to the 20,266 articles and 554 articles were returned. A total of 1,453 articles, published in 2007, were found when we performed a search using the keyword ‘cancer’ in the HuGE Literature Finder, an online curated tool (based solely on PubMed) that captures all articles on genetic associations, interactions and clinical and population studies of genetic tests and information (for details on the HuGE Navigator see Yu et al. [23]). Additionally, 1,293 unique results were returned when we performed a PubMed search for cancer related publications, published in 2007 involving human subjects. In this search, the keyword ‘gene’ was combined with one of the following keywords: behavioral and social sciences, communication, health services research, economics, surveillance, dissemination, diffusion and implementation.
We combined these 3 searches, removed duplicates and had a resulting 3,300 articles that were enriched for T2 and beyond. The same 5 reviewers who conducted the portfolio analysis read a total random 20% sample of the 3,300 collected abstracts for inclusion into our literature analysis (660 articles total). Reviewers categorized each relevant abstract by translational research phase (T0/T1/T2/T3/T4) and study type (basic/clinical/population). Abstracts categorized as T2 and beyond were further categorized as being either therapeutic or diagnostic. We then extrapolated the results back to the 20,266 cancer genetics publications that we found in our initial PubMed search. To do this we first extrapolated the 20% enriched sample that we read back to the entire 3,300 articles that were enriched for T2 and beyond. Next, we assumed that the 16,966 articles that did not appear in our enriched batch of articles were either T0 or T1 research. See figure 2 for a flow diagram of our methodology.
Methodology and results from the literature analysis of cancer genetics research articles, 2007.
Methodology and results from the literature analysis of cancer genetics research articles, 2007.
Results
NCI Portfolio Analysis
Of the 1,844 research grant abstracts that were identified based on our search criteria, 1,019 were deemed relevant for our analysis. Examples of types of studies in each phase are shown in table 1. As shown in figure 3a, 827 (81.16%) of these grants were classified as T0 and 174 (17.07%) were classified as T1. Only 18 (1.77%) of the grants we reviewed were classified as T2 and beyond: 9 were T2, 8 were T3 and 1 grant was classified as T4. Furthermore, as shown in figure 3b, of the grants classified as T0, 54% were considered basic science, 22% were clinical science and 24% were population-based. Of the 174 grants classified as T1, 2% were basic science, 70% were clinical science and 28% were population-based. Of the 9 T2 grants, 67% were clinical and 33% were population-based. Of the 8 T3 grants, 37% were clinical and 67% were population-based. The 1 T4 grant was population-based. Most T2 and beyond articles (89%) were coded as diagnostic and 11% were considered therapeutic studies.
Results of the National Cancer Institute cancer genetics research portfolio analysis, by translational research phase (a) and study type (b), Fiscal Year 2007.
Results of the National Cancer Institute cancer genetics research portfolio analysis, by translational research phase (a) and study type (b), Fiscal Year 2007.
Literature Review
After extrapolating the 3,300 cancer genetics publications from our literature review back to the 20,266 total cancer genetics publications from 2007, only 130 (0.64% of the 20,266) would have been considered T2 and beyond. Thirty-five articles were T2 research, 80 articles were T3 research and 15 articles were T4 research (fig. 2). Examples of types of studies in each phase are shown in table 1. Similar to the results of the NCI portfolio analysis, 89% of the T2 and beyond articles were coded as diagnostic and 11% were coded therapeutic. A good example of a validated cancer genetic application is BRCA1. The BRCA1 gene was discovered in 1994. Although BRCA1 testing was offered almost immediately by one company, translational research continued until the U.S. Preventive Services Task Force decided in 2005 that there was enough evidence to offer BRCA1 testing to women with high risk family history [12].
Discussion
In our systematic, cross-sectional investigation of translational research in cancer genetic research in 2007, we find that translational research in cancer genetics accounts for a very small fraction of the overall research portfolio in the field. These findings are consistent with genetic research in general and other fields of translational medicine [12]. They also clearly indicate the severe constriction in documented translational research after the initial phase of genetic discoveries and especially after the first phase of translation leading to a candidate application.
It is important to acknowledge the inherent strengths and limitations in this type of analysis. Because we relied on reading abstracts of funded NCI research, we acknowledge that these abstracts may reflect an incomplete picture of the scope of actual research that the investigators are performing. It is also important to note that the NCI grants do not reflect all of the cancer genetics research that is being performed (e.g., non-NCI funded research or the NCI intramural program). Finally, the portfolio analysis reflects currently funded cancer genetics research which could potentially predict future research publications. The literature analysis of 2007 reflects the status of publications in the field at a time when genome-wide association studies were just beginning to take off. We expect even greater numbers of genetic discovery publications in 2008 and beyond, thus increasing the numbers of T0 and T1 research. Another limitation is the potential under-ascertainment of T2 and beyond articles in the analysis of the literature. Because we did not read all 20,266 articles, we had to rely on a combination of approaches using medical subject headings, word searches and categories defined by the National Library of Medicine. Nevertheless, we believe our approach paints an overall picture of the status of translational research in cancer genetics in 2007 and of the severe constriction in research on candidate applications for cancer care and prevention. Furthermore, our duplicate coding and quality control review procedures ensured internal consistency in our definitions and classification of translational research phases.
What do these findings tell us and where should we go from here? Clearly, an appropriate emphasis on discovery research in cancer genetics is still needed since the amount and scope of large-scale biological, clinical and epidemiological information continues to emerge. Therefore, we should expect more discovery research in cancer genetics for years to come. Nevertheless, to reap the benefits of gene discoveries, there is a need to facilitate research and to build a research infrastructure that reflects translational clinical and population sciences for current and future discoveries. Such research translation requires a multidisciplinary approach. Even for well-known high penetrance genes that follow Mendelian inheritance patterns such as BRCA1 and Lynch syndrome, which are associated with cancer, we know very little about best ways to implement diagnosis and management of patients and their relatives (T3 research). Furthermore, we have very little data from the real world about the impact of the introduction of these tests on the overall burden of these cancers in the population (T4 research) [24]. For the more common genetic variants associated with weak to moderate increases in cancer risk, we do not yet know how to assess the clinical implications of these variants, especially their added value compared to more conventional cancer risk predictions (e.g., the Gail model) [25].
It is difficult to assess accurately why the distribution of cancer genomic research is skewed away from translation. This may reflect a combination of factors, although we have no direct data to assess their relative importance. These factors include the relative dearth of investigator applications for funding, the preferences of peer-review panels and the absence of infrastructure to conduct large-scale population research in epidemiology and multidisciplinary research such as behavioral sciences. An additional but unmeasured factor is that such translational research could be sponsored by other federal agencies and industry. Additionally, it is possible that some downstream therapeutic and diagnostic research is being performed by industry and thus would not be reflected in such a portfolio analysis. However, since the literature analysis did not ascertain more translation papers, we feel that this is unlikely to explain the large translational research gap. Certainly, the perception that more discovery is needed before embarking on translational research among researchers and peer reviewers may contribute to the mismatch between discovery and translational research.
Despite the present lack of translational research, we are already seeing the emergence of genetic profiles for susceptibility to several cancers, including prostate (http://www.decodediagnostics.com/PC-general.php) and breast cancers (http://www.myriad.com/products/bracanalysis.php). Many such markers are viewed as candidate health applications for risk assessment and cancer prevention. Some are even sold directly to consumers (e.g., https://www.23andme.com/). As recommended in an NIH-CDC multidisciplinary workshop on personal genomics in December 2008 by Khoury et al. [26], a multidisciplinary translational research agenda is needed to move genetic applications into practice and to document clinical benefits, costs and harms through observational studies and clinical trials. Additional research in behavioral and social sciences is needed to assess how genetic information impacts patients, providers, family members and the population at large [27]. Communication sciences will play an important role in determining the best way to educate and inform individuals about rapidly changing genetic information. Finally, outcomes research in the health services settings will be critical in documenting the impact of these technologies in practice and surveillance research to document their impact on population health. It is important to consider that translational research categories T1 to T4 correspond to the phase of translation and not to the type of research conducted. For example, behavioral and communication research can be done in T1 and T2 even before a specific genomic application is ready for implementation, and later in T3 and T4 as part of implementation and measuring population health outcomes.
In a time of rapidly escalating healthcare costs, it is becoming absolutely essential to document the added value of genetic information in cancer control and treatment beyond traditional approaches that do not use genetic information. This is particularly needed in the field of pharmacogenomics, where promising genetic tests may be able to tailor cancer therapeutics to minimize side effects and maximize drug response. Although very few clinical trials have been conducted in pharmacogenomics, the results of such trials have far reaching effects. For example, the Children’s Oncology Group recently found through a clinical trial that alteration of IKZF1, a gene that encodes the lymphoid transcription factorIKAROS, is strongly associated with recurrence in pediatric acute lymphoblastic leukemia [28].
The chasm between discovery research and translational research in cancer genetics is contributing to our ‘evidence dilemma’ in genomic and personalized medicine [29]. A series of articles by Khoury et al. [29], Woodcock [30] and Hudson [31] recently discussed the widening gap between discoveries and practice in genomic medicine and the need for a more robust applied research infrastructure that includes observational studies and randomized clinical trials. Even for genomic applications currently in practice, we lack highly needed information on how validated genome technologies (such as HER2 testing) actually work in practice [32] and whether or not they lead to ‘real world’ net benefits in clinical outcomes. In fact, over the past 7 years, independent panels such as the U.S. Preventive Services Task Force and the EGAPP Working Group only made a few evidence-based recommendations for use of genomic applications in practice [33,34]. Two recent evidence reviews of promising genomics applications yielded inconclusive evidence of clinical utility for their use in practice [35,36].
Although it can be argued that not all genetic factors and gene-environment interactions related to cancer are currently known, it is important to enhance the current translational research infrastructure to assess emerging applications. As discussed by Califf and Ginsburg, this will require enhancing the clinical epidemiology capacity through multidisciplinary teams, integration of clinical, molecular databases and population records, use of interoperable electronic health records and centralized biobanking [37]. Furthermore, even though most genetic risk factors are not ready for translation into practice, translational research in the form of assessing impact of genetic information on behavior can and should be done in proof-of-principle investigations. An example of this type of study is the ‘multiplex’ project conducted by McBride et al. [27]. The goal of this project, an example of T3 research, is to evaluate the impact of information from multiple variants that can spur individuals to seek additional risk assessments (e.g., family history and behavioral risk assessments) and/or additional health services (e.g., well care visits), even though the genetic markers used for the study will not be ones that are used in clinical practice in the future.
For validated genetic applications already in use, it is important to evaluate how we are doing in practice and whether or not all segments of the population are benefiting from these applications. The ‘lost in translation’ phenomenon has been described in other fields of medicine [38], where most basic science discoveries do not make it into clinical applications, and those that do take a very long time to get there [39], contributing to exacerbations in health care costs and disparities in our population. In summary, this paper clearly documents the present lack of a robust translational research agenda in cancer genetics. A translational research infrastructure is urgently needed to fulfill the promise of gene discoveries for cancer care and prevention in the 21st century.
References
The findings and conclusions in this report are those of the authors and do not necessarily reflect the views of the Department of Health and Human Services.