Abstract
Background: Quiescin sulfhydryl oxidase 1 (QSOX1), which oxidizes sulfhydryl groups to form disulfide bonds in proteins, is found to be over-expressed in various pancreatic cancer cell lines and patients. QSOX1 promotes invasion of pancreatic cancer cells by activating MMP-2 and MMP-9. However, its regulatory mechanism remains largely undefined. Methods: Real-time PCR and Western blot were employed to detect the expression of QSOX1 in human pancreatic cancer cell lines under hypoxic condition. Luciferase reporter and ChIP assays were used to assess the regulation of QSOX1 by hypoxia-inducible factor 1 (HIF-1). Small interfering RNA (siRNA) was applied to knock down endogenous expression of QSOX1. Matrigel-coated invasion chamber essays were conducted to detect the invasion capacity of QSOX1-depleted cells. Results: Both hypoxia and hypoxia mimicking reagent up-regulated the expression of QSOX1 in human pancreatic cancer cell lines. Knockdown of HIF-1α eliminated hypoxia induced QSOX1 expression. HIF-1α was found directly bound to two hypoxia-response elements (HRE) of QSOX1 gene, both of which were required for HIF-1 induced QSOX1 expression. Moreover, QSOX1 silencing blocked hypoxia-induced pancreatic cancer cells invasion. Conclusion: QSOX1 is a direct target of HIF-1 and may contribute to hypoxia-induced pancreatic cancer cells invasion.
Introduction
Pancreatic cancer is one of the most lethal malignancies with a 5-year survival rate of only about 5% in the United States [1]. Due to its asymptomatic progression, the majority of patients present with advanced and unresectable disease because of extensive local spread or metastatic disease at presentation. Therapeutic approaches against the advanced disease have largely failed and more than 80% of pancreatic cancer patients still die within 2-8 months after onset [2]. For this reason, elucidation of the mechanisms driving invasion and metastasis of pancreatic cancer cells is a prime research focus, as it may lead to the development of novel therapeutic modalities.
Hypoxia is one of the fundamental biological phenomena that plays a key role in the development and aggressiveness of a wide variety of cancers including pancreatic cancer [3,4]. The homeostatic response to hypoxia is predominantly mediated by the transcription factor hypoxia-inducible factor (HIF)-1 which has been widely accepted to play critical roles in tumor invasion, metastasis, and treatment resistance [5,6]. HIF-1 is a heterodimer complex that consists of a HIF-1α subunit and a HIF-1β subunit. Under normoxic conditions, HIF-1α is hydroxylated and degraded rapidly through von Hippel-Lindau (VHL) mediated ubiquitin-proteasome pathway [7]. While under hypoxic conditions, HIF-1α becomes stabilized, is rapidly accumulated in cell, and dimerizes with HIF-1β, thus translocates into cellular nucleus and activates genes of the hypoxia response by binding to the hypoxic response element (HRE) of its target genes [7].
QSOX1 belongs to the family of FAD-dependent sulfhydryl oxidases. The biological function of QSOX1 is to oxidate sulfhydryl members during protein folding to generate disulfide bonds in proteins, thus ultimately reducing the transformation from oxygen to hydrogen peroxide [8,9,10]. Recently, QSOX1 has been identified over-expressed in pancreatic and breast cancers and plays an important role in pancreatic and breast cancer cells invasion via activating MMP-2 and MMP-9 [11,12,13,14]. However, the regulation mechanism of QSOX1 remains largely elucidated.
In this study, we found that QSOX1 is induced by hypoxic stimuli and identified that QSOX1 is a direct target of HIF-1. Our data also suggested that elevated QSOX1 contributed to hypoxia induced MMP-2 and MMP-9 activation and pancreatic cancer cells invasion.
Material and Methods
Cell Culture
The pancreatic cancer cell line PANC-1 and CFPAC-1 cells were purchased from The Cell Bank of Type Culture Collection of Chinese Academy of Sciences (CAS, Shanghai), and cultured in DMEM medium supplemented with 10% fetal bovine serum (FBS) (Gibco, CA, USA) as well as 100U/ml penicilin and 100µg/ml streptomycin. Cells were placed in a 5% CO2 and 95% air incubator (20% O2) at 37°C. For hypoxia treatment, cells were cultured in a hypoxic chamber flushed with 1% O2 and 5% CO2 with balance of 95% nitrogen at 37°C.
SiRNA and Transfection
Small interfering (si) RNA against HIF-1α were purchased from Dharmacon (USA), with the following sequences: siHIF-1α-1 GGACACAGAUUUAGACUUG, and siHIF-1α-2 GAUGGAAGCACUAGACAAA, respectively. A random siRNA sequence was selected as control siRNA. shRNA vectors from the Mission human shRNA hQSOX1 clone sets (Sigma Aldrich) were used with each construct code as follows: shQSOX1-1 (TRCN0000064183), shQSOX1-2 (TRCN-0000064186), and an appropriate control vector: Control (SHGLYNM_001004128). All transient transfections were performed using the transfection reagent Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions.
RNA isolation and Real-time PCR
Total RNAs from PANC-1 and CFPAC-1 cells were isolated by TRIzol reagent, and reverse transcriptions were performed with Takara RNA PCR kit following manufacturer's instructions. In order to quantify the transcripts of the interest genes, real-time PCR was performed using a SYBR Green Premix Ex Taq (Takara, Japan) on Light Cycler 480 (Roche, Switzerland). QSOX1 transcript expression was normalized to β-actin mRNA expression using the ΔCt method and the linearized ΔCt was used for comparative purposes. The primer sequences used are listed as follows:
Luciferase assays
The QSOX1 promoter region was amplified from a human PANC-1 genomic DNA template and cloned into pGL3 basic vector (Promega). QSOX1 mutants were generated by using a PCR mutagenesis kit (Toyobo). For luciferase reporter assays, cells were seeded in 24-well plates and transfected with the indicated plasmids. Cells were then harvested 48 h after transfection. Luciferase activities were measured and assessed using the Dual Luciferase Reporter Assay System (Promega, USA).
ChIP Assays
Chromatin immunoprecipitation (ChIP) assay kits were used (Upstate, USA). In short, cells were fixed with 1% formaldehyde to cross-link the proteins and DNA, after which DNA was sheared to fragments ranging from 200 to 800 bps by being sonicated in an ultrasound bath on ice. The chromatin was then incubated and precipitated with anti-IgG and anti-HIF-1α antibodies. The immunoprecipitated DNA fragments were detected using real-time PCR analysis.
Western Blot
Cells were harvested and lysed with RIPA buffer (50 mM Tris-HCl, pH 7.4, 100 mM DTT, 1% w/v SDS, 10% glycerol). After being centrifuged at 20 000 g for 10 min at 4°C, proteins in the supernatants were quantified and separated by SDS-PAGE electrophoresis. Proteins were then transferred to NC membranes for 2h. Membranes were then blocked with 5% fat-free milk and immunoblotted with indicated antibodies, followed by HRP-linked secondary antibodies. The signals were detected by Millipore SuperSignal® HRP Substrate kit according to manufacturer's instructions.
Cell invasion assay
Cell invasion capacity was assessed using 24-well inserts (Becton Dickinson Labware, USA) with 8-mm pores according to manufacturer's protocol. The membranes utilized were Matrigel-coated invasion chambers (BD Biosciences, USA) that were pre-hydrated in serum-free medium. After 24 h of incubation, the cells in the upper chamber were removed, and the cells were fixed in ice-cold methanol, stained with Wright-Giemsa solution (Polysciences, USA). Digital images were obtained from the membranes, and cell areas were selected using Scan Scope CS system (Aperio Technologies, USA). The invasion cells were quantified in five randomly selected fields in each membrane, and the average value was defined as an invasion index on three independent membranes.
Statistical Analysis
Data were shown as mean ± SEM. Student's t test was used for statistical significance to assess experimental differences. Differences between groups were analyzed using Student's t-test. A p value less than 0.05 was considered statistically significant. Statistical significance is displayed as * (P < 0.05), ** (P < 0.01) or *** (P < 0.001).
Results
QSOX1 was induced by hypoxia and hypoxia-mimicking reagent in pancreatic cancer cells
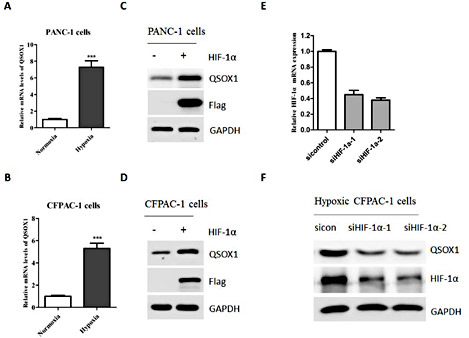
In order to determine whether QSOX1 expression is regulated by hypoxic stimuli, PANC-1 and CFPAC-1 cells were incubated under either normoxia (20% O2) or hypoxia (1% O2) for 24 h. The mRNA expression levels of QSOX1 in each group were detected by Real-time PCR. As shown in Fig. 1A, compared with those under normoxic condition, QSOX1 mRNA levels were significantly increased in the cells under hypoxic condition. Accordingly, QSOX1 protein levels were also enhanced in cells under hypoxic condition (Fig. 1B). PANC-1 cells treated with 100 μM hypoxia-mimetic reagent DFO (Deferoxamine mesylate salt) for 24 h also displayed an increase in QSOX1 protein expression (Fig. 1C). Furthermore, it was demonstrated that the hypoxia induced QSOX1 expression in PANC-1 cells in a time-dependent manner (Fig. 1D). Taken together, these data suggested that QSOX1 was induced by hypoxia in pancreatic cancer cells.
QSOX1 was induced by hypoxia and hypoxia-mimicking reagent in pancreatic cancer cells. A: Real-time PCR was employed to detect QSOX1 mRNA expression in PANC-1 cells under either normoxic or hypoxic conditions. B: Real-time PCR was used to detect QSOX1 mRNA expression in CFPAC-1 cells under either normoxic or hypoxic conditions. C: Lysates from PANC-1 cells under either normoxic or hypoxic conditions were detected with indicated antibodies, with GAPDH used as a loading control. D:Lysates from CFPAC-1 cells under either normoxic or hypoxic conditions were assessed with indicated antibodies. E: Lysates from PANC-1 cells treated with DMSO or DFO for 24h were detected with indicated antibodies. F: Lysates from PANC-1 cells under hypoxic condition for the indicated time courses were detected with indicated antibodies.
QSOX1 was induced by hypoxia and hypoxia-mimicking reagent in pancreatic cancer cells. A: Real-time PCR was employed to detect QSOX1 mRNA expression in PANC-1 cells under either normoxic or hypoxic conditions. B: Real-time PCR was used to detect QSOX1 mRNA expression in CFPAC-1 cells under either normoxic or hypoxic conditions. C: Lysates from PANC-1 cells under either normoxic or hypoxic conditions were detected with indicated antibodies, with GAPDH used as a loading control. D:Lysates from CFPAC-1 cells under either normoxic or hypoxic conditions were assessed with indicated antibodies. E: Lysates from PANC-1 cells treated with DMSO or DFO for 24h were detected with indicated antibodies. F: Lysates from PANC-1 cells under hypoxic condition for the indicated time courses were detected with indicated antibodies.
Transcriptional factor HIF-1 was required for QSOX1 expression during hypoxia
HIF-1 is a key transcriptional regulator in hypoxic response [15]. To test whether HIF-1 mediates hypoxia-regulated QSOX1 expression, HIF-1α plasmid was transfected into PANC-1 and CFPAC-1 cells for 48h. Notably, ectopic expression of HIF-1α could significantly up-regulate QSOX1 expression in both cell lines. To further investigate the role of HIF-1 in the regulation of QSOX1, PANC-1 cells were transfected with siRNAs targeting HIF-1α (siHIF-1α-1/2) to inhibit endogenous HIF-1α expression or non-specific gene (sicontrol) and thereafter incubated in hypoxic condition for 24h. The HIF-1α knockdown effect was evaluated by both real-time PCR and western blot (Fig. 2C and 2D), respectively. Consistent with the overexpression data, blocking the expression of HIF-1α by two different siRNAs significantly decreased hypoxia-induced QSOX1 expression (Fig. 2E and 2F).
Transcriptional factor HIF-1 was required for QSOX1 expression during hypoxia. A:PANC-1 cells were transfected with either an empty vector or HIF-1α plasmid for 48h. The mRNA levels of QSOX1 in both groups were detected by Real-time PCR. B: CFPAC-1 cells were transfected with either an empty vector or HIF-1α plasmid for 48h. The mRNA levels of QSOX1 in both groups were detected by Real-time PCR. C: PANC-1 cells were transfected with either an empty vector or HIF-1α plasmid for 48h. Both cells were harvested and the protein levels of QSOX1 were detected by Western blot. D: CFPAC-1 cells were transfected with either an empty vector or HIF-1α plasmid for 48h. Both cells were harvested and the protein levels of QSOX1 were detected by Western blot. E: CFPAC-1 cells were transfected with either con-siRNA or siRNAs against QSOX1 for 36h. The mRNA levels of QSOX1 were detected by Real-time PCR. F: CFPAC-1 cells were transfected with either con-siRNA or siRNAs against QSOX1 for 24h and incubated under hypoxic condition for additional 24h. Cells were harvested and the protein levels of QSOX1 were detected by Western blot.
Transcriptional factor HIF-1 was required for QSOX1 expression during hypoxia. A:PANC-1 cells were transfected with either an empty vector or HIF-1α plasmid for 48h. The mRNA levels of QSOX1 in both groups were detected by Real-time PCR. B: CFPAC-1 cells were transfected with either an empty vector or HIF-1α plasmid for 48h. The mRNA levels of QSOX1 in both groups were detected by Real-time PCR. C: PANC-1 cells were transfected with either an empty vector or HIF-1α plasmid for 48h. Both cells were harvested and the protein levels of QSOX1 were detected by Western blot. D: CFPAC-1 cells were transfected with either an empty vector or HIF-1α plasmid for 48h. Both cells were harvested and the protein levels of QSOX1 were detected by Western blot. E: CFPAC-1 cells were transfected with either con-siRNA or siRNAs against QSOX1 for 36h. The mRNA levels of QSOX1 were detected by Real-time PCR. F: CFPAC-1 cells were transfected with either con-siRNA or siRNAs against QSOX1 for 24h and incubated under hypoxic condition for additional 24h. Cells were harvested and the protein levels of QSOX1 were detected by Western blot.
Identification and validation of two functional HREs in QSOX1 promoter region
HIF-1 regulates target genes expression by binding to HIF-response elements (HREs) in promoters that contain the specific sequence 5'-NCGTG-3' [16]. By analyzing the QSOX1 gene sequence, we identified two putative HIF-1 response elements (HRE) respectively located at the first exon and the first intron region of QSOX1 gene (HRE1-28 5'- GCGTG -3'-23 and HRE2-801 5' -GCGTG- 3'-796) (Fig. 3A). We then generated a human QSOX1 promoter-driven luciferase reporter gene containing both HRE1 and HRE2. As expected, QSOX1 promoter activity was dramatically enhanced by hypoxic stimuli (Fig. 3B). Furthermore, mutation of either HRE1 or HRE2 largely lowered HIF-1α-induced luciferase gene expression and mutation of both HRE1 and HRE2 completely abolished HIF-1α-induced luciferase gene expression (Fig. 3C). In addition, chromatin immunoprecipitation assays in PANC-1 cells revealed that HIF-1α could bound to both HRE1 and HRE2 under hypoxic condition (Fig. 3D). Collectively, these foundings suggested that HIF-1's binding to the hypoxia response elements on the QSOX1 promoter was essentially required for QSOX1 induction.
Identification and validation of two functional HREs in QSOX1 promoter region. A:Schematic diagrams of the regulating sequences with the two putative HREs (black ovals) of QSOX1. B:CFPAC-1 cells were transfected with the putative QSOX1 HRE-luciferase reporter plasmid. 24 h after transfection, cells were cultured under indicated conditions for additional 24 h, after which the luciferase activities in each group were measured and compared, respectively. Transfection efficiency was normalized by Renilla luciferase expression. C: Schematic diagrams of the mutations within the two putative HREs of QSOX1. CFPAC-1 cells were co-transfected with or without HIF-1α plasmid and these WT or Mutant QSOX1 HRE-luciferase reporter plasmids. After 48 h of transfection, respective luciferase activity was measured and assessed. D: Anti-IgG and anti-HIF-1α antibodies were employed in the ChIP assays using CFPAC-1 cells incubated under either normoxic or hypoxic conditions.
Identification and validation of two functional HREs in QSOX1 promoter region. A:Schematic diagrams of the regulating sequences with the two putative HREs (black ovals) of QSOX1. B:CFPAC-1 cells were transfected with the putative QSOX1 HRE-luciferase reporter plasmid. 24 h after transfection, cells were cultured under indicated conditions for additional 24 h, after which the luciferase activities in each group were measured and compared, respectively. Transfection efficiency was normalized by Renilla luciferase expression. C: Schematic diagrams of the mutations within the two putative HREs of QSOX1. CFPAC-1 cells were co-transfected with or without HIF-1α plasmid and these WT or Mutant QSOX1 HRE-luciferase reporter plasmids. After 48 h of transfection, respective luciferase activity was measured and assessed. D: Anti-IgG and anti-HIF-1α antibodies were employed in the ChIP assays using CFPAC-1 cells incubated under either normoxic or hypoxic conditions.
QSOX1 contributed to hypoxia-induced pancreatic cancer cells invasion
It is well documented that hypoxia could trigger cancer cells invasion [17]. QSOX1 has been indicated to play an important role in pancreatic cancer invasion. To investigate whether QSOX1 has a role in hypoxia-induced pancreatic cancer invasion, we inhibited the expression of QSOX1 in hypoxically treated PANC-1 cells. Both shRNAs could efficiently knock down endogenous QSOX1 expression (Fig. 4A). By employing an invasion model, we confirmed that hypoxia could significantly promote PANC-1 cells' invasion capacity(Fig. 4B). However, QSOX1-depleted hypoxically cultured PANC-1 cells demonstrated a decreased invasive phenotype (Fig. 4C). A similar phenomenon was also observed in QSOX1-depleted hypoxically treated CFPAC-1 cells (Fig. 4D). Taken together, these results indicated that QSOX1 played an important role in hypoxia-induced pancreatic cancer cells invasion.
QSOX1 contributed to hypoxia-induced pancreatic cancer cells invasion. A: PANC-1 cells were transfected with indicated shRNAs for 48h. The protein levels of QSOX1 were then detected by Western blot. B: Invasion assays in PANC-1 cells under either normoxic or hypoxic conditions. C: Invasion assays in PANC-1 cells transfected with indicated shRNAs for 24h and incubated under hypoxic condition for additional 24h. D: Invasion assays in CFPAC-1 cells transfected with indicated shRNAs for 24h and incubated under hypoxic condition for additional 24h.
QSOX1 contributed to hypoxia-induced pancreatic cancer cells invasion. A: PANC-1 cells were transfected with indicated shRNAs for 48h. The protein levels of QSOX1 were then detected by Western blot. B: Invasion assays in PANC-1 cells under either normoxic or hypoxic conditions. C: Invasion assays in PANC-1 cells transfected with indicated shRNAs for 24h and incubated under hypoxic condition for additional 24h. D: Invasion assays in CFPAC-1 cells transfected with indicated shRNAs for 24h and incubated under hypoxic condition for additional 24h.
Discussion
In the present study we showed, for the first time to our knowledge, that hypoxia and hypoxia-mimicking reagent could significantly increase both the mRNA and protein levels of QSOX1 in human pancreatic cancer cells. Then by scanning the 5'-promoter region of QSOX1 for the consensus sequence of the HIF-1 response element (HRE), two potential binding sites for HIF-1 were revealed. By using luciferase and ChIP assays, we further proved that QSOX1 was a direct target gene for HIF-1 and both identified HREs were required for HIF-1α binding.
QSOX1 has been reported to be over-expressed in pancreatic cancer cell lines and pancreatic cancer samples and play an important role in pancreatic cancer invasion [11]. It has been suggested that in pancreatic tumor cells, QSOX1 directly contributes to an invasive and potentially metastatic phenotype partially through the activation of MMP2 and MMP9 [11]. However, how QSOX1 regulates the activity of MMP2 and MMP9 is completely unknown. The biological function of QSOX1 is to oxidate sulfhydryl members during protein folding to generate disulfide bonds in proteins, which ultimately reduce the shift from oxygen to hydrogen peroxide. However, whether the FAD-dependent sulfhydryl oxidase activity of QSOX1 is required for its invasion-promoting function remains completely elusive. The direct target of QSOX1 has not been yet identified, thus calling for further and more specific research and investigations. During the preparation of this work, a recently published paper showed that QSOX1 activity was required for the incorporation of laminin into the extracellular matrix (ECM) synthesized by fibroblasts, and ECM produced without QSOX1 was defective in supporting cell-matrix adhesion, which enriched our knowledge on QSOX1's biological roles [18]. Nevertheless, our data showed that the invasion ability of QSOX1-depleted hypoxically treated pancreatic cancer cells was dramatically decreased when compared with that of control-depleted cells. Thus, our data suggested an indispensable role of QSOX1 in hypoxia-induced pancreatic cancer invasion and suggested that QSOX1 could be a potential drug target for pancreatic cancer invasion and would be invaluable for guiding therapy.
Conflict of Interest
The authors declare that they have no conflict of Interest.